Molecular characterization of cultivated and wild genotypes of brinjal (Solanum melongena L.) using microsatellite markers
*Article not assigned to an issue yet
Thakare Vrunda S., Undirwade D. B., Khelurkar V. C., Moharil M. P., Ghawade S. M., Kulkarni U. S.
Research Articles | Published: 26 May, 2025
First Page: 0
Last Page: 0
Views: 535
Keywords: Genetic diversity, Microsatellite marker, Brinjal, Heterozygosity, EST-SSR
Abstract
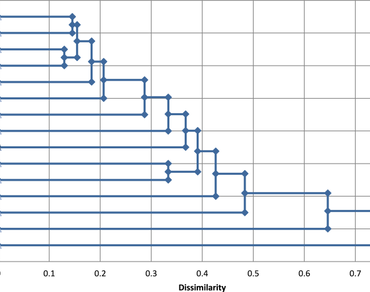
Genetic diversity has a paramount role in crop improvement and provide plants with capacity to meet the demands of changing environments. Brinjal is one of the important vegetable crops in India and Asian countries, but the information on genetic diversity of brinjal genotypes and wild relatives is rather limited compared to other solanaceous crops. With this view, present experiment was carried out to assess the diversity and extent of genetic relatedness among 14 local cultivated brinjal genotypes and one wild relative (S. viarum) for development of potential variety. The selected brinjal genotypes were characterized using 22 microsatellite loci (12 Inter Simple Sequence Repeat (ISSR) and 10 Expressed Sequence Tag-derived-Simple Sequence Repeat (EST-SSR)), distributed uniformly throughout the genome. The dendrogram was constructed based on un-weighted pair group method with arithmetic mean (UPGMA) to determine marker based genetic relationship amongst the selected genotypes. Computation of data was performed using the program XLSTAT software (www.xlstat.com). These 22 microsatellite loci amplified total of 116 alleles among 15 genotypes including wild relative with one to seven allele per locus with an average of 3.72. Highest polymorphism information content (PIC value) was observed to be 0.70 for marker EM 114. The observed polymorphism percentage ranged from 50 to 100 per cent, and heterozygosity ranged from 0.1 to 0.9 was detected across the selected genotypes. Polymorphism information content (PIC) ranged from 0.01 to 0.59 with a mean of 0.4, indicating high level of polymorphism across the selected genotypes. Jaccard’s dissimilarity coefficients between the cultivars, ranged from 0.324 to 0.966 with an average of 0.741. Cluster analysis (UPGMA) separated the selected genotypes into seven sub-groups, wild relative in separate cluster. This indicates the possible sharing of common genomic regions occurring across the genotypes. Comparatively, higher number of alleles, high polymorphism percentage, PIC values and observed heterozygosity were detected among the local genotypes compared with wild relative indicating rich genetic diversity among them. In view of these results, the local cultivated genotypes had significant genetic diversity along with one wild relative and constitute interesting genotypes which become promising material for potential breeding program.
References
Adeniji OT, Kusolwa PM, Reuben SOWM (2012) Genetic diversity among accessions of Solanum aethiopicum L. groups based on morpho-agronomic traits. Plant GeNet Resour: Characterisation and Utilisation 10:177–185
Adjebeng-Danquah J, Gracen VE, Offei SK et al (2016) Agronomic performance and genotypic diversity for morphological traits among cassava genotypes in the guinea savannah ecology of Ghana. J Crop Sci Biotechnol 19:99–108
Aguoru CU, Omoigui LO, Olasan JO (2015) Population genetic study of eggplants (Solanum) species in Nigeria, Tropical West Africa, using molecular markers. Int J Plant Res 5(1):7–12
Ahmed F, Rabbani MG, Islam MR et al (2019a) Molecular characterization of some brinjal genotypes (Solanum melongena L) using simple sequence repeat (SSR) markers. Afr J Agric Res 14:1980–1989
Aina OO, Dixon AGO, Paul I, Akinrinde EA (2009) GxE interaction effects on yield and yield components of cassava (landraces and improved) genotypes in the savanna regions of Nigeria. Afr J Biotech 8:4933–4945
Anushma PL, Rajasekharan PE, Sing TH (2018) A review on availability, utilization and future of eggplant genetic resources in India. J Plant Dev Sci 10(12):645–657
Balyejusa KE, Chiwona-Karltun L, Egwang T et al (2006) Genetic diversity and variety composition of cassava on small-scale farms in Uganda: an interdisciplinary study using genetic markers and farmer interviews. Genetica 130(3):301–318
Bindler G, Vander Hoeven R, Gunduz I et al (2006) A microsatellite marker based linkage map of tobacco. Theor Appl Genet 114(2):341–349
Blair MW, Mccouch SR, Panaud O (1999) Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor Appl Genet 98(5):780–792
Chavarriaga-Aguirre P, Maya MM, Bonierbale MW et al (1998) Microsatellites in Cassava (Manihot esculenta Crantz): discovery, inheritance and variability. Theor Appl Genet 97(3):493–501
Demir K, Bakir M, Sarikamis G, Acunalp S (2010) Genetic diversity of eggplant (Solanum melongena) germplasm from Turkey assessed by SSR and RAPD markers. Genet Mol Res : GMR 9:1568–1576
Diez MJ, Nuez F (2008) Tomato. In: Prohens J, Nuez F (eds) Handbook of plant breeding: vegetables II. Springer publication, New York, pp 249–323
Ferguson ME, Hearne SJ, Close TJ et al (2011) Identification, validation and high-throughput genotyping of transcribed gene SNPs in cassava. Theor Appl Genet 124(4):685–695
Fu YB (2006) Impact of plant breeding on genetic diversity of agricultural crops: searching for molecular evidence. Plant Genetic Resources 4:71–78
Islam Z, Siddiqua MK, Hasan MM et al (2014) Assessment of genetic diversity of brinjal (Solanum melongena L.) germplasm by RAPD markers. Research on Crops 15:416–422
Kaur A, Kaur N, Bassi G et al (2020) Morphogical and molecular markers based assessment of genetic diversity in eggplant. Indian J Hortic 77:116–124
Mangal M, Upadhyay P, Kalia P (2016) Characterization of cultivated and wild genotypes of brinjal (Solanum melongena L.) and confirmation of hybridity using microsatellite markers. Vegetos 29(2):8
Nunome T, Suwabe K, Iketani H et al (2003) Identification and characterization of microsatellites in eggplant. Plant Breed 122:256–262
Nunome T, Negoro S, Kono I et al (2009) Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theor Appl Genet 119(6):1143–1153
Ojuderie OB, Balogun MO, Fawole I et al (2015) Assessment of the genetic diversity of African yam bean (Sphenostylis stenocarpa Hochst ex. A Rich. Harms) accessions using amplified fragment length polymorphism (AFLP) markers. Afr J Biotech. https://doi.org/10.5897/AJB2014.13734
Pattanaik A, Reddy LD, Singh TH et al (2018) Optimization of sampling size for DNA-based PCR assay for hybrid purity test in the brinjal Solanum melongena. Biosci Biotechnol Res Commun 11(3):496–504
Rotino GL, Sala T, and Toppino, L (2014) Eggplant. Alien gene transfer in crop plants, volume 2: Achievements and impacts, 381–409.
Saini DK, Kaushik P (2019) Visiting eggplant from a biotechnological perspective: a review. Sci Hortic 253:327–340
Salgon S, Jorda C, Sauvage C et al (2017) Eggplant resistance to the Ralstonia solanacearum species complex involves both broad-spectrum and strain-specific quantitative trait loci. Front Plant Sci 8:254776
Sharma AD, Gill PK, Singh P (2002) DNA isolation from dry and fresh samples of polysaccharide-rich plants. Plant Mol Biol Report 20:415–415
Stagel A, Portis E, Toppino L et al (2008) Gene-based microsatellite development for mapping and phylogeny studies in eggplant. BMC Genomics 9:1–14
Sultana S, Islam MN, Hoque ME (2018) Dna fingerprinting and molecular diversity analysis for the improvement of brinjal (Solanum melongena l.) cultivars. J Adv Biotechnol Exp Ther 1:01–06
Syfert MM, Castaneda-Alvarez NP, Khoury CK et al (2016) Crop wild relatives of the brinjal eggplant (Solanum melongena): Poorly represented in genebanks and many species at risk of extinction. Am J Bot 103:635–651
Yi G, Lee JM, Lee S et al (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet 114(1):113–130
Author Information
Department of Agricultural Entomology, PGI, Dr. PDKV Akola, Akola, India