Identification and characterization of GSTase gene family in different developmental stages of C. cajan using various software and computational tools of plant genome biology
*Article not assigned to an issue yet
Singh Nootan, Sahu Sumanta Kumar, Kondabala Rajesh, Vaish Swati
Research Article | Published: 19 August, 2025
First Page: 0
Last Page: 0
Views: 31
Keywords: n Cajanus Cajann , Glutathione S-transferase, Expression profiles, Legume information system (LIS)
Abstract
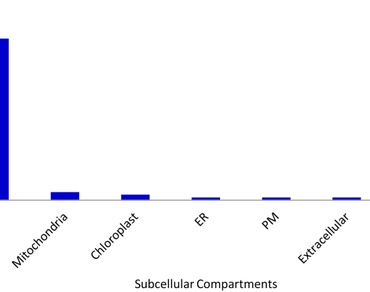
Plant glutathione S-transferases (GSTs) are multifunctional conserved protein superfamily that is involved in various biological processes such as growth and development, cellular detoxification, stress biology, and various signaling processes. In the current study, a comprehensive genome-wide identification and characterization of the GST gene family were performed in the agriculturally important legume crop Cajanus cajan. A total of 68 GST genes were identified that belong to eight GST classes based on their conserved domains and motifs. Among 68 CcGST genes, 37 CcGST genes were found on seven Cajanus chromosomes and the remaining CcGST genes were found on the scaffold. Both segmental and tandem duplications were key drivers of CcGST gene family expansion. The conserved pattern of exon and intron structure among the different GST classes was observed. The physicochemical properties of major CcGST proteins reveal that they are acidic in nature. The expression profiling study revealed the high expression of CcGSTU38, CcGSTU40, CcGSTU44, CcGSTL3, CcGSTL4, CcEF1G1, CcEF1G2, CcDHAR2 and CcGSTF6 in most of the developmental stages in different anatomical tissues. The molecular docking and MD simulation study of highly expressed CcGSTU38 with the herbicide safeners revealed its highest binding affinity and structural stability with Fenclorim (-5.44 kcal/mol). This gene could be a potential candidate for future molecular characterization under herbicide stress. The results of the current study endow us with the further functional analysis of Cajanus GSTs in the future.
References
Abramson J, Adler J, Dunger J et al (2024) Accurate structure prediction of biomolecular interactions with alphafold 3. Nature 630:493–500. https://doi.org/10.1038/s41586-024-07487-w
Abraham M, Alekseenko A, Bergh C, Blau C, Briand E, Doijade M, Fleischmann S et al (2023) ‘GROMACS 2023.3 Source Code’. Zenodo. https://doi.org/10.5281/zenodo.10017686
Armenteros JJA, Sonderby CK, Sonderby SK, Nielsen H, Winther O (2017) DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 21:3387–4339. https://doi.org/10.1093/bioin
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 37(suppl_2):W202–W8. https://doi.org/10.1093/nar/gkp335 PMID: 19458158
Cao Q, Lv W, Jiang H, Chen X, Wang X, Wang Y (2022) Genome-wide identification of glutathione S-transferase gene family members in tea plant (Camellia sinensis) and their response to environmental stress. Int J Biol Macromol 205:749–760. https://doi.org/10.1016/j.ijbiomac.2022.03.109
Chen CJ, Chen H, He YH, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. https://doi.org/10.1101/289660. BioRxiv
Chronopoulou E, Georgakis N, Nianiou-Obeidat I, Madesis P, Perperopoulou F, Pouliou F et al (2017) Plant glutathione transferases in abiotic stress response and herbicide resistance. Glutathione in plant growth, development, and stress tolerance. Springer, London, pp 215–233
Martins DS, Vasquez LS, Tillack AF, Sanner MF, Koch A, Forli S (2021) J Chem Theory Comput 17:1060–1073. https://doi.org/10.1021/acs.jctc.0c01006
Dixon DP, Edwards R (2010) Glutathione transferases. Arabidopsis Book 8:e0131. https://doi.org/10.1199/tab.0131
Dong Y, Li C, Zhang Y, He Q, Daud MK, Chen J et al (2016) Glutathione S-transferase gene family in Gossypium raimondii and G. arboreum: Comparative genomic study and their expression under salt stress. Front in Plant Sci 7:139. https://doi.org/10.3389/fpls. 2016. 00139
Edwards R, Del Buono D, Fordham M, Skipsey M, Brazier M, Dixon DP, Cummings I (2005) Differential induction of glutathione transferases and glucosyltransferases in wheat, maize and Arabidopsis Thaliana by herbicide safeners. Z Naturforsch C J Biosci 60(3–4):307–316. https://doi.org/10.1515/znc-2005-3-416
FAO Statistics (2017) Pigeon producing countries. Production and area harvested. Food and Agriculture Organization of the United Nations, Rome
Fang X, An Y, Zheng J, Shangguan L, Wang L (2020) Genome-wide identification and comparative analysis of GST gene family in Apple (Malus domestica) and their expressions under ALA treatment. 3 Biotech 10(7):307. https://doi.org/10.1007/s13205-020-02299-x
Fiser A (2004) Template-based protein structure modeling. Methods Mol Biol 673:73–94. https://doi.org/10.1007/978-1-60761-842-3_6
Gao J, Chen B, Lin H, Liu Y, Wei Y, Chen F, Li W (2020) Identification and characterization of the glutathione S-transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene 743:144484. https://doi.org/10.1016/j.gene.2020.144484
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel RD et al (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook 571–607.https://doi.org/10.1385/1-59259-890-0:571
Ghangal R, Rajkumar MS, Garg R et al (2020) Genome-wide analysis of glutathione S-transferase gene family in Chickpea suggests its role during seed development and abiotic stress. Mol Biol Rep 47:2749–2761. https://doi.org/10.1007/s11033-020-05377-8
González-Faune P, Sánchez-Arévalo I, Sarkar S, Majhi K, Bandopadhyay R, Cabrera-Barjas G, Gómez A, Banerjee A (2021) Computational study on temperature driven structure–function relationship of polysaccharide producing bacterial Glycosyl transferase enzyme. Polymers 13:1771. https://doi.org/10.3390/polym13111771
Hasan MS, Islam S, Hasan MN, Sajib SD, Ahmed S, Islam T et al (2020) Genome-wide analysis and transcript profiling identify several abiotic and biotic stress-responsive glutathione S-transferase genes in soybean. Plant Gene 23:100239. https://doi.org/10.1016/j.plgene.2020.100239
Hasan MS, Singh V, Islam S, Islam MS, Ahsan R et al (2021) Genome wide identification and expression profiling of glutathione S-transferase family under multiple abiotic and biotic stresses in Medicago truncatula L. PLoS ONE 16:e0247170. https://doi.org/10.1371/journ. pone. 02471 70
Holub EB (2001) The arms race is ancient history in arabidopsis, the wildflower. Nat Rev Genet 2:516–527. https://doi.org/10.1038/35080
Horton P, Park K, Obayashi T, Fujita N, Harada H, Adams-Collier CJ et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587 (Web server issue). https://doi.org/10.1093/nar/gkm259
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Jain M, Ghanashyam C, Bhattacharjee A (2010) Comprehensive expression analysis suggests overlapping and specific roles of glutathione S-transferases during development and stressresponses in rice. BMC Genomics 11:73. https://doi.org/10.1186/1471-2164-11-73
Kayum MA, Nath UK, Park JI, Biswas MK, Choi EK, Song JY, Kim HT, Nou IS (2018) Genome-wide identifcation, characterization, and expression profiling of glutathione S-transferase (GST) family in pumpkin reveals likely role in cold-stress tolerance. Genes 9(2):84. https://doi.org/10.3390/genes9020084
Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in arabidopsis, arabis, and related genera (Brassicaceae). Mol Biologyand Evol 17(10):1483–1498. https://doi.org/10.1093/oxfordjournals.molbev.a026248
Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D (2013) Genome wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom 14:433. https://doi.org/10.1186/1471-2164-14-433
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Labrou NE, Papageorgiou AC, Pavli O, Flemetakis E (2015) Plant GSTome: structure and functional role in xenome network and plant stress response. Current Op in Biotechnol 32:186–194. https://doi.org/10.1016/j.copbio
Lallement PA, Brouwer B, Keech O, Hecker A, Rouhier N (2014) The still mysterious roles of cysteine-containing glutathione transferases in plants. Front Pharmacol 5:192. https://doi.org/10.3389/fphar.2014.00192
Letunic I, Bork P (2020) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46(D1):D493–D496. https://doi.org/10.1093/nar/gkaa937
Liu YJ, Han XM, Ren LL, Yang HL, Zeng QY (2013) Functional divergence of the glutathione S-transferase supergene family in physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol 161:773–786.
Marchler Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S et al (2017) CDD/SPARCLE: functional classifcation of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Mohanta TK, Khan A, Hashem A et al (2019) The molecular mass and isoelectric point of plant proteomes. BMC Genomics 20:631. https://doi.org/10.1186/s12864-019-5983-8
Pal D, Mishra P, Sachan N, Ghosh AK (2011) Biological activities and medicinal properties of Cajanus Cajan (L) Millsp. J Adv Pharm Technol Res 2(4):207–214. https://doi.org/10.4103/2231-4040.90874
Panchy N, Lehti-Shiu M, Shiu SH (2016) Evolution of gene duplication in plants. Plant Physiol 171(4):2294–2316. https://doi.org/10.1104/pp.16.00523
Pandey P, Pandey V, Kumar R, Tiwari D (2013) Imperative quality factors which influence nutritional value of pigeonpea [(Cajanus cajan (l.) millsp.)]. Agri-environment: perspectives on sustainable development, 169
Rastogi S, Liberles DA (2005) Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol 14:5:28. https://doi.org/10.1186/1471-2148-5-28
Shao D, Li Y, Zhu Q, Zhang X, Liu F, Xue F, Sun J (2021) GhGSTF12, a glutathione S-transferase gene, is essential for anthocyanin accumulation in cotton (Gossypium hirsutum L). Plant Sci 305:110827. https://doi.org/10.1016/j.plantsci.2021.110827
Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Song W, Zhou F, Shan C, Zhang Q, Ning M, Liu X, Zhao X, Cai W, Yang X, Hao G, Tang F (2021) Identification of glutathione S-transferase genes in Hami Melon (Cucumis Melo var. saccharinus) and their expression analysis under cold stress. Front Plant Sci 12(672017). https://doi.org/10.3389/fpls.2021.672017
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. https://doi.org/10.1093/nar/gkl315
Vaish S, Awasthi P, Tiwari S, Tiwari SK, Basantani MK (2018) In silico genome-wide identification and characterization of glutathione S-transferase gene family in Vigna radiata (L.) Wilczek. Genome 61:311–322. https://doi.org/10.1139/gen- 2017 – 0192
Vaish S, Gupta D, Basantani MK (2020) Glutathione S-transferase: a versatile protein family. 3 Biotech 10:321. https://doi.org/10.1007/s13205-020-02312-3
Vaish S, Parveen R, Gupta D, Basantani MK (2022) Genome-wide identification and characterization of glutathione S-transferase gene family in Musa acuminata L. AAA group and gaining an insight to their role in banana fruit development. J Appl Genet Doi. https://doi.org/10.1007/s13353-022-00707-x
Varshney R, Chen W, Li Y et al (2012) Draft genome sequence of Pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89. https://doi.org/10.1038/nbt.2022
Wang R, Ma J, Zhang Q et al (2019) Genome-wide identification and expression profling of glutathione transferase gene family under multiple stresses and hormone treatments in wheat (Triticum aestivum L). BMC Genomics 20:986. https://doi.org/10.1186/s12864-019-6374-x
Author Information
Varanasi, Uttar Pradesh, India