Genetic analysis and characterization of diverse pigeonpea genotypes for yield-enhancing traits
PS Basavaraj, KM Boraiah, Gupta Purvi, Raskar Nikhil, Rane Jagadish
Research Articles | Published: 06 September, 2023
First Page: 1627
Last Page: 1636
Views: 3075
Keywords: Diversity, Variability, Heritability, Correlation, Path analysis, Grain legumes, Pulses
Abstract
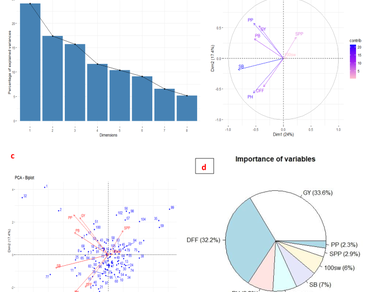
Developing improved pigeonpea varieties is vital to ensure food and nutritional security. However, understanding the nature and extent of genetic variability and trait association is crucial to design an appropriate breeding strategy. Hence, the present study aimed to ascertain the genetic variability through multivariate analysis in a diverse pigeonpea germplasm for yield-enhancing traits. Traits, number of primary branches per plant (NPB), secondary branches per plant (NSB), pods per plant (PP), and grain yield per plant (GY) possessed high variability. High heritability coupled with high genetic advance was recorded for days to 50% flowering (DFF), NPB, NSB, PP, test weight (TW), and GY. Trait association revealed grain yield is significantly and positively associated with PH, NPB, NSB, PP, SPP, and TW. Path analysis revealed that the highest direct positive effect of traits on grain yield was by PP followed by NSB, NPB, SPP, and TW. Principal component analysis revealed that the first two PCs accounted for maximum variability and explained 42.01% of the total variability. The traits, PP and SPP contributed the maximum to variance. Thus, the selection of genotypes with a higher number of PP and higher SPP would be rewarding. Cluster analysis grouped 106 genotypes into ten clusters. Selection of the genotypes from different clusters based on their performance would yield superior recombinants upon hybridization. Overall, the study revealed the pattern of genetic variability present in a set of pigeonpea genotypes for yield-enhancing traits and this scientific lead helps in designing appropriate breeding strategies for the genetic improvement of pigeonpea cultivars.
References
Basavaraj PS, Gireesh C, Muralidhara B, Manoj CA, Anantha MS, Damodar Raju CH (2020) Genetic analysis of backcross derived lines of Oryza rufipogon in the background of Samba Mahsuri a for yield-enhancing traits in rice. Electron J of Plant Breed 11(04):1120–1127
Basavaraj PS, Gireesh C, Bharamappanavara M, Manoj CA, Ishwarya Laxmi VG, Honnappa, Ajitha V, Senguttuvel P, Sundaram RM, Anantha MS (2021a) Stress tolerance indices for the identification of low phosphorus tolerant introgression lines derived from Oryza rufipogon Griff. Plant Genetic Resources: Characterization and Utilization 1–11. https://doi.org/10.1017/S1479262121000381
Basavaraj PS, Muralidhara B, Manoj CA, Anantha MS, Santosha Rathod DR, Ch, Senguttuvel P, Madhav MS, Srinivasaprasad M, Prakasam V, Basavaraj K, Jyothi Badri, Subbarao LV, Sundaram RM, Gireesh C (2021b) Identification and molecular characterization of high-yielding, blast resistant lines derived from Oryza rufipogon Griff. In the background of ‘Samba Mahsuri’ rice. Genet Resour Crop Evol 68:1905–1921. https://doi.org/10.1007/s10722-020-01104-1
Basavaraj PS, Gireesh C, Bharamappanavara M, Manoj CA, Ishwaryalakshmi LVG, Senguttuvel P, Sundaram RM, Subbarao LV, Anantha MS (2022) Genetic analysis of introgression lines of Oryza rufipogon for improvement of low phosphorous tolerance in indica rice. Indian J Genet Plant Breed 82(02):135–142. https://doi.org/10.31742/IJGPB.82.2.1
Bhandari HR, Bhanu AN, Srivastava K, Singh MN, Shreya Hemantaranjan A (2017) Assessment of genetic diversity in crop plants - an overview. Adv Plants Agric Res 7:279–286. https://doi.org/10.15406/apar.2017.07.00255
Bohra A, Saxena KB, Varshney RK, Saxena RK (2020) Genomics-assisted breeding for pigeonpea improvement. Theor Appl Genet 133(5):1721–1737. https://doi.org/10.1007/s00122-020-03563-7
Fufa H, Baenziger PS, Beecher BS et al (2005) Genetic improvement trends in agronomic performances and end-use quality characteristics among hard red winter wheat cultivars in Nebraska. Euphytica 144:187–198. https://doi.org/10.1007/s10681-005-5811-x
Hemavathy AT, Kannan Bapu JR, Priyadarshini C (2017) Principal component analysis in Pigeonpea (Cajanus cajan (L.)Millsp). Electron J Plant Breed 8(4):1133–1139. https://doi.org/10.5958/0975-928X.2017.00165.X
India-Stat (2021) https://www.indiastat.com/data/agriculture/total-foodgrains/data-year/2022
Johnson HW, Robinson HF, Comstock RE (1955) Estimates of genetic and environmental variability of soybeans. Agron J 47:314–318
Jolliffe IT, Cadima J (2016) Principal component analysis: a review and recent developments. Phil Trans R Soc A. https://doi.org/10.1098/rsta.2015.0202
Kandarkar K, Kute NS, Tajane S, Anant I (2020) Correlation and path analysis for yield and its contributing traits in pigeonpea [Cajanus cajan (L.) Mill Sp]. J Pharmacognosy Phytochemistry 9(4):1629–1632
Li R, Li M, Ashraf U, Liu S, Zhang J (2019) Exploring the relationships between yield and yield-related traits for rice varieties released in China from 1978 to 2017. Front Plant Sci 10:543
Mehmood S, Bashir A, Ahmad A, Akram Z, Jabeen N, Gulfraz M (2008) Molecular characterization of regional Sorghum bicolor varieties from Pakistan. Pakistan J of Bot 40:2015–2021
Muniswamy S, Lokesha R, Dharmaraj PS, Yamanura, Diwan JR (2014) Morphological characterization and assessment of genetic diversity in minicore collection of pigeonpea [Cajanus Cajan (L.) Millsp]. Electron J Plant Breed 5(2):179–186
Rekha RM, Prasanthi R, Sekhar, Shanti priya M (2013) Principal component and cluster analyses in pigeonpea [Cajanus cajan (L.) Millsp]. Int J Applied Biology and Pharmaceutical Technol 4(4):424–430
Renwick LLR, Kimaro AA, Hafner JM, Rosenstock TS, Gaudin ACM (2020) Maize-pigeonpea intercropping outperforms Monocultures under Drought. Front sustain food sys 4:562663. https://doi.org/10.3389/fsufs.2020.562663
Sandeep S, Sujatha K, Chitturi MM, Sudha Rani C (2020) Genetic analysis of pigeonpea [Cajanus cajan (L.) Millsp.] Hybrids for yield and yield attributes. Legume Res - Inter J. https://doi.org/10.18805/LR-4280
Shukla S, Mishra SP, Mehandi S (2022) Exploiting the natural variation and inter-relationship among medium maturity pigeonpea [Cajanus cajan (L.) Millsp.] Genotypes. Electron J Plant Breed 13(2):717–723
Susmitha D, Kalaimagal T, Senthil R, Vetriventhan M, Manonmani S, Jeyakumar P, Anita B, Reddymalla S, Choudhari PL, Nimje CA, Peerzada OH, Arveti VN, Azevedo VCR, Singh K (2022) Grain nutrients variability in Pigeonpea Genebank Collection and its potential for promoting Nutritional Security in Dryland Ecologies. Front Plant Sci 13:934296. https://doi.org/10.3389/fpls.2022.934296
Varthini VN, Robin S, Sudhakar D, Raveendran M, Rajeswari S, Manonmani S (2014) Evaluation of rice genetic diversity and variability in a population panel by principal component analysis. Indian J Science and Tech 7(10):1555–1562
Won PLP, Liu H, Banayo NPM, Nie L, Peng S, Islam MR, Cruz PS, Collard BCY, Kato Y (2020) Identification and characterization of high-yielding, short-duration rice genotypes for tropical Asia. Crop Sci 60:2241
Zavinon F, Adoukonou-Sagbadja F, Bossikponnon A, Dossa H, Ahanhanzo C (2019) Phenotypic diversity for agro-morphological traits in pigeonpea landraces (Cajanus cajan L.) Millsp.) Cultivated in southern Benin. Open Agric 4:487–499. https://doi.org/10.1515/opag-2019-0046
Author Information
ICAR-National Institute of Abiotic Stress Management, Baramati, India