Gene expression and survival analysis study of KIAA0101 gene revealed its prognostic and diagnostic importance in breast cancer
Research Articles | Published: 13 January, 2023
First Page: 249
Last Page: 258
Views: 3546
Keywords: Breast cancer, Microarray, KIAA0101 , Welch t test, Survival analysis, Kaplan–Meier plot
Abstract
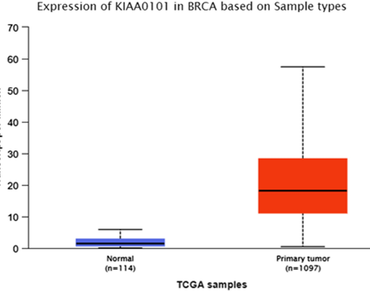
Breast cancer (BC) is the most frequent cancer affecting women worldwide. High-throughput genomic platforms and microarray-based gene expression profiling have emerged as one of the best strategies for understanding and interpreting BC at the genetic level. BRCA1/2 are the main target gene while several other genes also play crucial role in BC. In the present study, we evaluated the expression, diagnostic and survival/prognostic role of the KIAA0101 gene in BC. We applied the student t test (equal or unequal sample sizes, unequal variances/Welch’s t test) and the Kaplan–Meier estimator to validate dependability of the KIAA0101 (PCLAF, PCNA clamp associated factor) gene in BC. Microarray data (GSE10810) were retrieved from “Gene Expression Omnibus”, “National Center for Biotechnology Information (NCBI)”. KIAA0101 gene was one of the significantly expressed genes while comparing 31 breast tumor samples with 27 normal breast samples (control) at the threshold (fold change of ± 2 and p value < 0.001) using the Welch t test. The Kaplan–Meier Plotter tool was used to verify the overall survival and the relapse free survival of the KIAA0101 gene on bigger and 50 independent GEO datasets; 20 BC, 13 ovarian cancer, 11 lung cancer, and 6 gastric cancer cohorts. Hazard ratio and log rank p value of the KIAA0101 gene were determined under different restrictions of estrogen receptors, progesterone receptors and lymph node status (positive and negative). Finally, the wFisher, Lancaster, and weighted Z-method robust meta-analysis and statistical approaches were used to combine p-values for collective statistical significance of KIAA0101 gene. Score of log rank p value (< 0.05) indicated the therapeutic and prognostic importance of gene. Furthermore, to improve the precision of the analysis, qPCR was done to validate microarray expression result of KIAA0101 gene. In conclusion, KIAA0101 was one of the most over-expressed genes in BC which can be considered as a potential prognostic and diagnostic biomarker.
References
De Biasio A, de Opakua A, Mortuza G, Molina R, Cordeiro T, Castillo F, Villate M, Merino N, Delgado S, Gil-Cartón D, Luque I, Diercks T, Bernadó P, Montoya G, Blanco F (2015) Structure of p15PAF–PCNA complex and implications for clamp sliding during DNA replication and repair. Nat Commun 6(1):6439
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S (2022) UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 25:18–27. https://doi.org/10.1016/j.neo.2022.01.001. (PMID: 35078134)
Delacre M, Lakens D, Leys C (2017) Why psychologists should by default use Welch’s t-test instead of Student’s t-test. Int Rev Soc Psychol 30(1):92–101. https://doi.org/10.5334/irsp.82
Dwivedi AK, Mallawaarachchi I, Alvarado LA (2017) Analysis of small sample size studies using nonparametric bootstrap test with pooled resampling method. Stat Med 36(14):2187–2205. https://doi.org/10.1002/sim.7263. (Epub 2017 Mar 9, PMID: 28276584)
Erceg-Hurn D, Mirosevich V (2008) Modern robust statistical methods: an easy way to maximize the accuracy and power of your research. Am Psychol 63(7):591–601. https://doi.org/10.1037/0003-066X.63.7.591
Evangelou E, Ioannidis JP (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 14(6):379–389. https://doi.org/10.1038/nrg3472
Fan S, Li X, Tie L, Pan Y, Li X (2015) KIAA0101 is associated with human renal cell carcinoma proliferation and migration induced by erythropoietin. Oncotarget 7(12):13520–13537
He Y, Cao Y, Wang X, Jisiguleng W, Tao M, Liu J, Wang F, Chao L, Wang W, Li P, Fu H, Xing W, Zhu Z, Huan Y, Yuan H (2021) Identification of hub genes to regulate breast cancer spinal metastases by bioinformatics analyses. Comput Math Methods Med 2021:1–12
Kais Z, Barsky S, Mathsyaraja H, Zha A, Ransburgh D, He G, Pilarski R, Shapiro C, Huang K, Parvin J (2011) KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol Cancer Res 9(8):1091–1099
Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M (2012) Overexpression of KIAA0101 predicts poor prognosis in primary lung cancer patients. Lung Cancer 75(1):110–118
Lánczky A, Győrffy B (2021) Web-based survival analysis tool tailored for medical research (KMplot): development and Implementation. J Med Internet Res 23(7):e27633. https://doi.org/10.2196/27633
Liu J, He Y, Li C, Zhou R, Yuan Q, Hou J, Wu G (2021) Increased KIAA0101 gene expression associated with poor prognosis in breast cancer. Transl Cancer Res 10(9):4009–4019
Lv W, Su B, Li Y, Geng C, Chen N (2018) KIAA0101 inhibition suppresses cell proliferation and cell cycle progression by promoting the interaction between p53 and Sp1 in breast cancer. Biochem Biophys Res Commun 503(2):600–606. https://doi.org/10.1016/j.bbrc.2018.06.046
Pedraza V, Gomez-Capilla J, Escaramis G, Gomez C, Torné P, Rivera J, Gil A, Araque P, Olea N, Estivill X, Fárez-Vidal M (2010) Gene expression signatures in breast cancer distinguish phenotype characteristics, histologic subtypes, and tumor invasiveness. Cancer 116(2):486–496. https://doi.org/10.1002/cncr.24805
Petroziello J, Yamane A, Westendorf L, Thompson M, McDonagh C, Cerveny C, Law C, Wahl A, Carter P (2004) Suppression subtractive hybridization and expression profiling identifies a unique set of genes overexpressed in non-small-cell lung cancer. Oncogene 23(46):7734–7745
Tantiwetrueangdet A, Panvichian R, Sornmayura P, Leelaudomlipi S, Macoska JA (2021) PCNA-associated factor (KIAA0101/PCLAF) overexpression and gene copy number alterations in hepatocellular carcinoma tissues—BMC Cancer. BioMed Cent. https://doi.org/10.1186/s12885-021-07994-3
Tseng GC, Ghosh D, Feingold E (2012) Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res 40(9):3785–3799. https://doi.org/10.1093/nar/gkr1265
Whitlock MC (2005) Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J Evol Biol 18(5):1368–1373
Yoon S, Baik B, Park T, Nam D (2021) Powerful P-value combination methods to detect incomplete association. Sci Rep. https://doi.org/10.1038/s41598-021-86465-y
Author Information
University Department of Statistics and Computer Applications, Tilka Manjhi Bhagalpur University, Bhagalpur, India