Extremely diverse structural organization in the complete mitochondrial genome of seedless Phoenix dactylifera L
Research Articles | Published: 04 March, 2019
First Page: 92
Last Page: 97
Views: 3872
Keywords: Mitochondrial genome, Phoenix dactylifera , Seedless mutant, Nucleotide variations, SNPs
Abstract
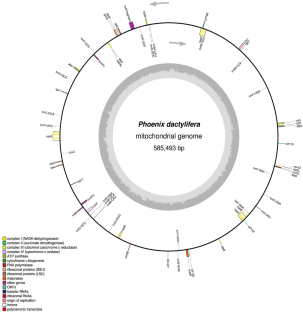
Mitochondrial genome of seedless (mutant) variety of Phoenix dactylifera is analyzed to understand the probable reason for seedless condition. The size of the genome is 585,493 bp long which is 129,508 bp shorter compared to the normal genome and demonstrate significantly higher rate of nucleotide loss. The loss is evident in coding and non-coding regions. Missing of all rRNA genes and other three genes, i.e., rps14, rps13, rps19 is observed. Total 2726 SNPs are recorded, showed 80.9% identity with the normal genome. The ratio of nonsynonymous (dN) synonymous (dS) substitution (dN/dS) is more than one for nad7, nad1, ccMb, ccmC, cob, matR, matB suggest adaptive selection while less than one for cox2 suggest strong selective pressure on this gene and reduction in size may be start of its transfer to nucleus. Such variations may play an important role in seedless condition is discussed.
References
- Al-Khalifah N, Askari E (2003) Molecular phylogeny of date palm (Phoenix dactylifera L.) cultivars from Saudi Arabia by DNA fingerprinting. Theor Appl Genet 107:1266–1270
- Al-Shahib W, Marshall RJ (2003) The fruit of the date palm: its possible use as the best food for the future? Int J Food Sci Nutr 54:247–259
- Baliga MS, Baliga BRV, Kandathil SM, Bhat HP, Vayalil PK (2011) A review of the chemistry and pharmacology of the date fruits (Phoenix dactylifera L.). Food Res Int 44:1812–1822
- Bock R (2010) The give-and-take of DNA: horizontal gene transfer in plants. Plant Sci 15:11–22
- Chen Z et al (2017) Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species. Plant Biol 19:484–493
- Cohen Y, Korchinsky R, Tripler E (2004) Flower abnormalities cause abnormal fruit setting in tissue culture-propagated date palm (Phoenix dactylifera L.). J Hortic Sci Biotechnol 79:1007–1013
- Cuenca A, Ross TG, Graham SW, Barrett CF, Davis JI, Seberg O, Petersen G (2016) Localized retroprocessing as a model of intron loss in the plant mitochondrial genome. Genome Biol Evol 8:2176–2189
- Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
- Galtier N (2011) The intriguing evolutionary dynamics of plant mitochondrial DNA. BMC Biol 9:61
- Gray MW (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4:a011403
- Gray MW et al (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res 26:865–878
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283:1476–1481
- Grewe F, Edger PP, Keren I, Sultan L, Pires JC, Ostersetzer-Biran O, Mower JP (2014) Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 19:135–143
- Gualberto JM, Newton KJ (2017) Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol 68:225–252
- Gugerli F, Sperisen C, Büchler U, Brunner I, Brodbeck S, Palmer JD, Qiu Y-L (2001) The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol Phylogenet Evol 21:167–175
- Hepburn NJ, Schmidt DW, Mower JP (2012) Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol Biol Evol 29:3111–3120
- Joly S, Brouillet L, Bruneau A (2001) Phylogenetic implications of the multiple losses of the mitochondrial coxII.i3 intron in the angiosperms. Int J Plant Sci 162:359–373
- Kitazaki K, Kubo T (2010) Cost of having the largest mitochondrial genome: evolutionary mechanism of plant mitochondrial genome. J Bot 2010:12
- Kudla J, Albertazzi F, Blazević D, Hermann M, Bock R (2002) Loss of the mitochondrial cox2 intron 1 in a family of monocotyledonous plants and utilization of mitochondrial intron sequences for the construction of a nuclear intron. Mol Genet Genomics 267:223–230
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
- Larkin MA et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
- Liu Y, Xue J-Y, Wang B, Li L, Qiu Y-L (2011) The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution. PLoS One 6:e25836
- Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575–W581
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD (2007) Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7:135
- Mower JP, Sloan DB, Alverson AJ (2012) Plant mitochondrial genome diversity: the genomics revolution. In: Wendel JF, Greilhuber J, Dolezel J, Leitch IJ (eds) Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. Springer, Vienna, pp 123–144
- Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu Y-L, Song K (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci 97:6960–6966
- Petersen G et al (2017) Mitochondrial genome evolution in Alismatales: size reduction and extensive loss of ribosomal protein genes. PLoS One 12:e0177606
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
- Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T (2004) The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics 272:247–256
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z (2012) An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One 7:e31468
- Sloan DB, Müller K, McCauley DE, Taylor DR, Štorchová H (2012) Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol 196:1228–1239
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8:35–46
- Tang M et al (2015) Rapid evolutionary divergence of Gossypium barbadense and G. hirsutum mitochondrial genomes. BMC genomics 16:770
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11
- Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci 84:9054–9058
- Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17:32–43
- Zhang Z, Li J, Yu J (2006) Computing Ka and Ks with a consideration of unequal transitional substitutions. BMC Evol Biol 6:44
Author Information
Department of Biosciences, Saurashtra University, Rajkot, India