Assessment of genetic diversity among species mungo-radiata group of subgenus Ceratotropis of genus Vigna Savi. using amplified fragment length polymorphism (AFLP)
Research Articles | Published: 17 August, 2023
First Page: 1443
Last Page: 1456
Views: 3071
Keywords: AFLP, Asiatic Vignan , Ceratotropis, Population substructure, Gene flow
Abstract
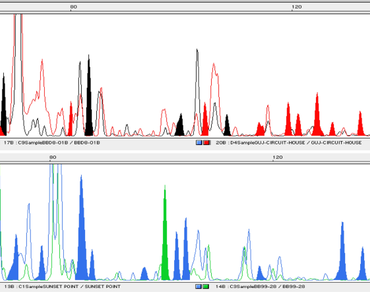
Genetic diversity, population substructure, species relationships, and gene flow was evaluated in 85 accessions belonging to the mungo-radiata group of section Ceratotropis of subgenus Ceratotropis of genus Vigna using amplified fragment length polymorphism. Twelve preselected AFLP primers generated 1869 polymorphic amplification products. The number of fragments for each primer pair ranged from 44 to 296, showing 100% polymorphism. An analysis of the mungo-radiata group from different phytogeographical regions across India revealed a high level of genetic diversity. The six species of the mungo-radiata group were assembled into three groups namely, Radiata, Mungo, and Hainiana. The highest total diversity as well as within accession diversity was obtained for the Hainiana group and minimum for the Mungo group. Genetic differentiation values for Mungo and Radiata group was 0.158 and 0.212, respectively. A high gene flow was detected within each group. For mungo-radiata-hainiana relatives, Fsc, Fct, and Fst estimates were 0.120, 0.089, and 0.034, respectively, and all Vigna species combined, the fixation indices Fst, Fsc and Fct were 0.120, 0.068, and 0.034, respectively. A tree with a star-shaped topology was obtained, in which the accessions from all the species are intermingled further substantiates gene flow between populations.
References
Arens P, Coops H, Jansen J, Vosman B (1998) Molecular genetic analysis of black poplar (Populous nigra) along Dutch rivers. Mol Ecol 7:11–18
Beebe S, Toro Ch, Gonzalez O, Chacon MI, Debouck DG (1997) Wild-weed-crop groups of common bean (Phaseolus vulgaris L., Fabaceae) in the Andes of Peru and Columbia, and their implications for conservation and breeding. Genet Res Crop Evol 44:73–91
Bhat KV, Lakhanpaul S, Chadha S (2005) Amplified fragment length polymorphism (AFLP) analysis of genetic diversity in Indian mungbean [Vigna radiata (L.) Wilczek] cultivars. Indian J Biotechnol 4:56–64
Bhattacharyya P, Ghosh S, Mandi SS et al (2017) Genetic variability and association of AFLP markers with some important biochemical traits in Dendrobium thyrsiflorum, a threatened medicinal orchid. S Afr J Bot 109:214–222
Brunk CF, Jones KC, James TW (1979) Assay for nanogram quantities of DNA in cellular homogenates. Annal Biochem 92:497–500
Egawa Y, Nakagawara M, Fernandez GCJ, Gatehouse AMR (1990) Cross compatibility and cytogenetical relationships among Asian Vigna species. In: Fuzii K, Johnson CD, Mitchel R, Yoshida T (eds) Bruchids and legumes: economics, ecology and coevolution. Kluwer Academic Publishers, pp 201–208
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction sites. Genetics 131:479–491
Gaiotto FA, Bramucci M, Grattapaglia D (1997) Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers. Theor Appl Genet 95:842–849
Govindraju DR (1988) Relationship between dispersal ability and levels of gene flow in plants. Oikos 52:31–35
Harlan JR (1965) The possible role of weed races in the evolution of cultivated plants. Euphytica 14:173–176
Harlan JR, DeWet JMJ (1965) Some thoughts about weeds. Econ Bot 19:16–24
Jaccard P (1908) Etude comparative de a distribution florale dans portiondes Alpes et des Jura. Bull Soc Vaud Sci Nat 37:547–579
Kaga A, Tomooka N, Egawa Y, Hosaka K, Kamijima O (1996) Species relationships in the subgenus Ceratotropis (genus Vigna) as revealed by RAPD analysis. Euphytica 88:17–24
Karp A, Seberg O, Buiatti (1996) Molecular techniques in the assessment of botanical diversity. Ann Bot 78:143–149
Koopman WJ, Zevenbergen MJ, Van den Berg RG (2001) Species relationships in Lactuca sativa. (Lactuceae, Asteraceae) inferred from AFLP fingerprints. Am J Bot 88(10):1881–1887
Lewontin R (1972) The apportionment of human diversity. Evol Biol 6:381–398
Liersch A, Bocianowski J, Popławska W et al (2019) Creation of gene pools with amplified fragment length polymorphism markers for development of winter oilseed rape (Brassica napus L.) hybrid cultivars. Euphytica 215:22
Majer D, Mithen R, Lewis BG, Vos P, Oliver RP (1996) The use of AFLP fingerprinting for the detection of genetic variation in fungi. Mycol Res 100:1107–1111
McDermott JM, McDonald BA (1993) Gene flow among plant pathosystems. Annu Rev Phytopathol 31:353–373
Misra A, Shassany AK, Shukla AK, Darokar MP, Singh SC, Sundaresan V, Singh J, Bagchi GD, Jain SP, Saikia D, Khanuja SP (2010) AFLP markers for identification of Swertia species (Gentianaceae). Genet Mol Res 9:1535–1544
Mueller UG, Wolfenbarger LL (1999) AFLP genotyping and fingerprinting. TREE 14:389–394
Muluvi GM, Sprent JI, Soranjo N, Provan J, Odee D, Folkard G, McNicol JW, Powell W (1999) Amplified Fragment Length Polymorphism (AFLP) analysis of genetic variation in Moringa oleifera Lam. Mol Ecol 8:463–470
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5673
Oka HI, Morishima H (1971) The dynamics of plant domestication: cultivation experiments with Oryza perennis and its hybrid with O. sativa. Evolution 25:356–364
Oliveira TG, Pereira AMS, Coppede JS et al (2016) Genetic diversity analysis of Croton antisyphilIiticus Mart. using AFLP molecular markers. Genet Mol Res 15(1):1–8
Paun O, Schönswetter P (2012) Amplified fragment length polymorphism: an invaluable fingerprinting technique for genomic, transcriptomic, and epigenetic studies. Methods Mol Biol 862:75–87
Rohlf FJ (2002). NTSYS-pc Numerical taxonomy and multivariate taxonomy system, version 2.11, Exeter software, Seteauket, New York
Saravanakumar P, Kaga A, Tomooka N, Vaughan DA (2004) AFLP and RAPD analyses of intra-and interspecific variation in some Vigna subgenus Ceratotropis (Leguminosae) species. Aust J Bot 52:417–424
Seehalak W, Tomooka N, Waranyuwar A et al (2006) Genetic diversity of the Vigna germplasm from Thailand and neighboring regions revealed by AFLP analysis. Genet Resour Crop Evol 53:1043–1059
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Sheeja TE, Kumar IPV, Giridhari A, Minoo D, Rajesh MK, Babu KN (2021) Amplified fragment length polymorphism: applications and recent developments. Methods Mol Biol 2222:187–218. https://doi.org/10.1007/978-1-0716-0997-2_12. (PMID: 33301096)
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39(1):53–65
Smartt J (1980) Evolution and evolutionary problems in food legumes. Econ Bot 34:219–235
Soltis PS, Bloom WL (1986) Genetic variation and estimates of gene flow in Clarkia speciosa subsp. polyantha (Onagraceae). Am J Bot 73(12):1677–1782
Tomooka N, Yoon MS, Doi K, Kaga A, Vaughan D (2002) AFLP analysis of diploid species in the genus Vigna subgenus Ceratotropis. Genet Res Crop Evol. 49:521–530
Tripathi N, Saini N, Tiwari S (2011) Assessment of genetic diversity among Aloe vera accessions using amplified fragment length polymorphism. Int J Med Arom Plants 1:115–121
Tripathi N, Saini N, Nair P, Tiwari S (2012) Lack of genetic diversity of a critically endangered important medicinal plant Chlorophytum borivilianum in central India revealed by AFLP markers. Physiol Mol Biol Plants 18(2):161–167
Varma A, Shrivastava N (2018) Genetic structuring in wild populations of two important medicinal plant species as inferred from AFLP markers. Plant Biosyst 152(5):1088–1100
Vir R, Bhat KV, Lakhanpaul S (2009) Analysis of population substructure, genetic differentiation and phylogenetic relationships among selected Asiatic Vigna species. Genet Res Crop Evol 56:783–795
Vos P, Hogers R, Bleeker R, Reijans M, VandeLee T, HomesM FA, Pot P, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nuc Acids Res 23:4407–4414
Wanjala BW, Obonyo M, Wachira FN, Muchugi A, Mulaa M, Skilton RA, Proud J, Hanson J (2013) Genetic diversity in Napier grass (Pennisetum purpureum) cultivars: implications for breeding and conservation. AoB Plants. 5:plt022
Author Information
Department of Botany, Zakir Husain Delhi College, Delhi University, New Delhi, India