Phylogenetic analysis of selected Asiatic Vigna species based on PCR–RFLP of three non-coding cpDNA Loci
Research Articles | Published: 01 November, 2022
First Page: 1172
Last Page: 1179
Views: 4276
Keywords: PCR–RFLP, Genetic diversity, Ceratotropis , TrnLUAA intron, TrnLUAA–trnFGAA and psbA-trnHGUG
Abstract
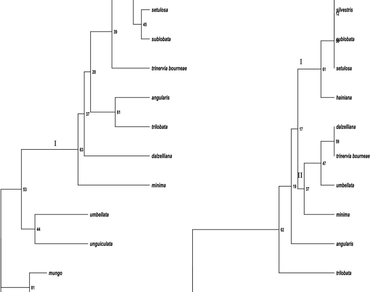
The PCR–RFLP was used to analyze the genetic diversity and relationship between cultivated species and wild taxa belonging to subgenus Ceratotropis of the genus Vigna. The chloroplast DNA was analyzed by amplification followed by restriction of three non-coding regions, trnLUAA intron, trnLUAA–trnFGAA and psbA-trnHGUG spacers. The length of intergenic spacers trnLUAA–trnFGAA and psbA-trnHGUG varied from 475 to 580 bp. The length of trnLUAA intron varied from 600 to 700 bp. Wild forms V. radiata var. sublobata and V. mungo var. silvestris emerged as distinct taxa and grouped close to their proposed cultivated forms viz. V. radiata and V. mungo. V. radiata var. setulosa, another wild form grouped closer to V. radiata var. sublobata and V. radiata. More importantly, V. hainiana appeared to be closely related to V. mungo and V. radiata, therefore needs to be investigated for its role in the domestication of these two important pulse yielding taxa.
References
Babu CR et al (1985) Taxonomic revision of Indian Phaseolus, Vigna, Macroptilium and Dysolobium. Bull Bot Surv Ind 27:1–28
Bisht IS et al (2004) Diversity and genetic resources of wild Vigna species in India. Genetic resources and crop evolution (in press)
Bukhari YM et al (1999) Phylogenetic analysis of Acacia (Mimosceae) as revealed from chloroplast RFLP data. Theor Appl Genet 98:291–298
Chandel KPS (1984) The wild ancestors of urid and mung bean (V. mungo (L.) Hepper and V. radiata (L.) Wilczek). Bot J Linn Soc 89:85–96
Cipriani G, Testolin R, Gardener R (1998) Restriction site variation of PCR-amplified chloroplast DNA regions and its implications for the evolution and taxonomy of Actinidia. Theor Appl Genet 96:389–396
Demesure B et al (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Demesure B et al (1996) Chloroplast DNA phylogeography of the common beech (Fagus sylvatica L.) in Europe. Evolution 50:2515–2520
Dumolin S et al (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256
Dumolin-Lapegue S et al (1997) An enlarge set of consensus primers for the study of organelle DNA in plants. Mol Ecol 6:393–397
Egawa Y et al (1996). Collaborative research program on mungbean germplasm (subgenus Ceratotropis of genus Vigna) between DOA Thailand and JIRCAS, Tsukuba, Japan. In: Mungbean germplasm: collection evaluation and utilization for breeding program, edited by Srinives P, Kitbamroong C and Miyazaki S, JIRCAS, Tsukuba, Japan, pp 1–8
El Mousadik A, Petit RJ (1996) Chloroplast DNA phylogeography of the argan tree of Morocco. Mol Ecol 5:547–555
Fineschi S et al (2000) Chloroplast DNA polymorphism reveals little geographic structure in Castanea sativa Mill. (Fagaceae) throughout Southern European countries. Mol Ecol 9:1495–1503
Frey WM et al (1999) Chloroplast DNA-relationship in palaeoaustral Lopidium concinnum (hypopterygiaceae) an example of stenoevolution in Moses. Studies in Austral temperate rainforest bryophytes 2. Plant Syst Evol 218:67–75
Harris SA, Ingram R (1991) Chloroplast DNA and biosystematics: the effects of intraspecific diversity and plastid transmission. Taxon 40:393–412
Jaaska V, Jaaska V (1990) Isozyme variation in Asian beans. Bot Acta 103:281–290
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kaga A et al (1996) Species relationships in the subgenus Ceratotropis (genus Vigna) as revealed by RAPD analysis. Euphytica 88:17–24
Kim SC et al (1999) The use of non-coding region of chloroplast DNA in phylogenetic studies of the subtribe Sonchinae (Asteraceae: Lactuceae). Plant Syst Evol 215:85–99
Mohanty A, Martin JP, Aguinagalde (2001) A population genetic analysis of chloroplast DNA in wild population of Prunus avium L. in Europe. Heredity 87:421–427
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5673
Ogihara Y, Terachi T, Sasakuma T (1991) molecular analysis of the hot spot region related to the length mutation in wheat chloroplast DNAs. I nucleotide divergence of genes and intergenic spacer regions located in the hotspot region. Genetics 129:321–332
Palmer JD (1987) Chloroplast DNA evolution and biosystematic uses of chloroplast DNA. Am Nat 130:S6–S29
Parani M et al (2000) Molecular phylogeny of mangroves VII. PCR–RFLP of trnS-psbC and rbcL gene regions in 24 mangrove-associated species. 100:454–460
Santalla M et al (1998) Genetic diversity in mungbean germplasm revealed by RAPD markers. Plant Breed 117:473–478
Taberlet P et al (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tateishi Y (1985) A revision of Azuki bean group, the subgenus Ceratotropis of genus Vigna (Leguminoseae) Ph.D. Dissertation, Tohuku University, Japan
Tomooka N et al (2002) AFLP analysis of diploid species in the genus Vigna subgenus Ceratotropis. Genet Res Crop Evol 49:521–530
Tsumura Y et al (1995) Molecular phylogeny of conifers using RFLP analysis of PCR amplified specific chloroplast genes. Theor Appl Genet 91:1222–1232
Tsumura Y et al (1996) Molecular phylogeny of Dipterocarpaceae in Southeast Asia using RFLP of PCR amplified chloroplast genes. Theor Appl Genet 93:22–29
Doi K, Kaga A, Tomooka N, Vaughan DA (2002) Molecular phylogeny of genus Vigna subgenus Ceratotropis based on rDNA ITS and atpB-rbcL intergenic spacer for cpDNA sequences. Genetica 114:129–145
Ignacimuthu S, Babu CR (1987) Mutagenesis for resistance to storage pest in the wild and cultivated urd and mungbean. J Nucl Agric Biol 16:169–176
Goel S, Raina SN, Ogihara Y (2001) Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of nuclear ribosomal DNA in the Phaseolus–Vigna group. Mol Phylogenet Evol 22(1):1–19
Rohlf FJ (2002) NTSYS-pc Numerical taxonomy and multivariate taxonomy system, version 2.11, Exeter software, Seteauket, New York
Singh BV, Ahuja MR (1977) Phaseolus sublobatus Roxb. A source of resistance to yellow mosaic virus for cultivated mung. Indian J Genet 37:130–132
Tomooka N, Lairingreang C, Nakeeraks P, Egawa Y, Thavarasook C (1992) Center of genetic diversity and dissemination pathways in mungbean deduced from seed protein electrophoresis. Theor Appl Genet 83:289–293
Lawn RJ, Williams RW, Imrie BC (1988) Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean. In: Mungbean: proceedings of the second international symposium. AVRDC, Taiwan, pp 136–145
Miyazaki H (1982) Classification and phylogenetic relationship of the Vigna radiata–silvestris–sublobata group. Bull Nat Ints Sci D 33:1–61
Fuzi K, Miyazaki S (1987) Infestation resistance of wild legumes Vigna sublobata to azukibean weevil, Callosobruchus chinensis (L.) (Coleoptera: Bruchidaceae) and its relationship with cytogenetic classification. Appl Entomol Zool 22:229–230
Fuzii K, Ishimoto M, Kitamura K (1989) Patterns of resistance to bean weevils (Bruchidae) in Vigna radiata-mungosublobata complex inform the breeding of new resistant varieties. Appl Entomol Zool 24(1):126–132
Author Information
Zakir Husain Delhi College Department of Botany, Delhi University, Delhi, India