Isolation, molecular identification and antibiotics susceptibility of Salmonella serotypes isolates from bats in Osun State, Nigeria
Aladejana Oluwatoyin Modupe, Oluduro Anthonia Olufunke, Ogunlade Ayodele Oluwayemisi, Babatunde Shola Kola, Sanni Olufunke Tolulope
Research Articles | Published: 02 February, 2024
First Page: 668
Last Page: 675
Views: 2901
Keywords: Bats, Salmonella species, Profile Index 20E Kit, Random amplified polymorphic DNA, Antibiotics susceptibility
Abstract
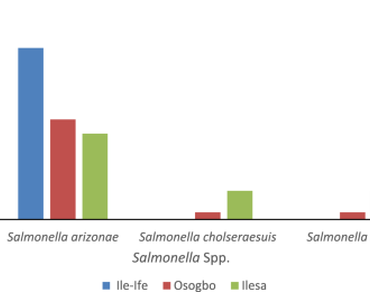
Bats (Chiroptera) are widely distributed in nature and are important reservoir host of many zoonotic bacterial pathogens such as Salmonella species. Bats faecal samples were collected from Ile-Ife, Osogbo and Ilesa and were streak cultured on Salmonella-Shigella agar and incubated aerobically at 37 °C. Non-lactose fermenting colonies were picked for further identification using biochemical tests and confirmation using Analytical Profile Index 20E Kit. The antibiotics susceptibility test on the confirmed Salmonella serotypes was performed using Kirby-Bauer’s disc diffusion technique. The degree of genetic relatedness and the phylogenetic diversity between the strains of the isolates were determined by random amplified polymorphic DNA (RAPD-PCR). Five different Salmonella spp were identified across the three locations; Salmonella arizonae, S. bongori, S. typhi, S. enterica, S. choleraesuis. S. arizonae was most abundant and common to the three locations (Ile-Ife, Osogbo and Ilesa). Resistance to antibiotics varied greatly among the isolates across the locations. All the isolates were susceptible to ciprofloxacin. Resistance to commonly available antibiotics such as gentamicin, oflaxacin, meropenem. eterpenem, sulphomethaxozole, tetracycline and nitrofuranton was significantly low (P ≥ 0.5) compared with other classes of antibiotics tested across the three locations. Resistance to augmentin (93.37%), cefuroxime (83.84%) ceftazidime (68.42%) were relatively high across the three locations. The random amplified polymorphic DNA revealed genetic polymorphism among the isolates and the strains were highly heterogeneous base on the distribution of different nucleotide sequences.
References
Abike TO, Osuntokun OT, Modupe AO, Adenike AF, Atinuke AR (2020) Antimicrobial efficacy, secondary metabolite constituents, ligand docking of Enantia chlorantha on selected multidrug resistance bacteria and fungi. J Adv Biol Biotechnol. 23(6):17–32. https://doi.org/10.9734/JABB/2020/v23i630161
Adesiyun AA, Stewart-Johnson A, Thompson NN (2009) Isolation of enteric pathogens from bats in Trinidad. J Wildl Dis 45(4):952–961
Akobi B, Aboderin O, Sasaki T, Shittu A (2012) Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU) Nigeria. BMC Microbiol 12:279
Aladejana OM, Oluduro AO (2021) Molecular characterization of extended spectrum β-lactamaseproducing- Enterobacteriaceae from bats (Eidolon helvum) faeces in Osun State Nigeria. Biologica Nyssana. 12(1):79–86. https://doi.org/10.5281/zenodo.5523057
Aladejana OM, Oluyege JO, Olowomofe TO, Obayemi IE, Oluyege DE (2021a) Multiple antibiotic resistance of airborne bacteria in outdoor markets in Ado-Ekiti metropolis. Serb J Exp Clin Res. https://doi.org/10.2478/sjecr-2020-0042
Aladejana OM, Famurewa O, Onyia CI, Thonda OA, Ogidi CO, Olawoye AA (2021b) Isolation and multiple antibiotic resistance pattern of enteric bacteria isolated from feacal samples of Thryonomys swinderianus in Sekona Osun State, Nigeria. Asian J Res Anim Vet Sci 7(4):23–28
Aladejana OM, Oluduro AO, Ogunlade AO, Famurewa O (2022) Molecular characterization of resistance and virulence genes in Escherichia coli isolated from bats (Eidolon helvum) faeces in Osun State Nigeria. J Adv Microbiol. https://doi.org/10.9734/jamb/2022/v22i230436
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259
Casset-Beraud AM, Richard C (1988) The aerobic intestinal flora of the microchiroptera bat Chaerephen pumila bats in Anjiro Madagascar. Archievers De Intstitut Pasteur, Madagascar 1989:233–239
CLSI (2018) Performance standards for antimicrobial susceptibility testing, Twentieth Informational Supplement, CLSI Document M100–S22. Clinical and Laboratory Standards Institute, Wayne
Costa DP, Poeta Y, Saenz L, Vinue AC, Coelho M, Matos B, Rojo-Bezares J, Rodrigues CT (2008) Mechanisms of Escherichia coli isolates recovered from wild animals. Microbiol Drug Resist 14:71–77
Costa PM, Loureiro L, Matos AJF (2013) Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Resourc Public Health 10:278–294
Di Bella C, Piraino CS, Caracappa L, Fornasari CV, Zara B (2003) Enteric microfloria in Itakan Chiroptera. Journal of Mt Ecology 7:221–224
Foley SL, Lynne AM (2008) Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci 86:173–187
Grimont PAD, Weill FX (2007) Antigenic formulae of the Salmonella Serovars, 9th ed., World Health Organization Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France
Mahajan RK, Khan SA, Chandel DS, Kumar N, Hans C, Chaudhry R (2003) Fatal case of Salmonella enterica subsp arizonae Gastroenteritis in an Infant with Microcephaly. J Clin Microbiol 41(12):5830–5832
Mereno G, Lopes CCA, Seabra E, Paran C, Correa A (1975) Bacteriological study of intestinal flora of bats (Desmodus rotundus). Arquivos Do Instituto Biologico (sao Paulo) 42:229–232
Michael CA, Dominey-Howes D, Labbate M (2014) The antibiotic resistance crisis: Causes, consequences and management. Front Public Health 2(145):00145
Olatimehin A, Shittu AO, Onwugamba FC, Mellmann A, Becker K, Schaumburg F (2018) Staphylococcus aureus complex in the Straw-Colored Fruit Bat (Eidolon helvum) in Nigeria. Front Microbiol 13(9):162. https://doi.org/10.3389/fmicb.2018.00162. (PMID: 29487577; PMCID: PMC5816944)
Oluduro AO (2012) Antibiotics resistant commensal Escherichia coli in faecal droplets from bats and poultry in Nigeria. Vet Ital 48(3):297–308
Osuntokun OT, Mayowa A, Thonda OA, Aladejana OM (2019) Pre/post plasmid curing and killing kinetic reactivity of discorea bulbifera linn against multiple antibiotics resistant clinical isolates using Escherichia coli as a case study. Int J Cell Sci Mol Biol. https://doi.org/10.19080/IJCSMB.2019.06.555685
Reed KD, Meece JK, Henkel JS, Shukla SK (2003) Birds, migration and emerging zoonoses: West nile virus, lyme disease, influenza A and enteropathogens. Clin Med Res 1:5–12
Santos L, Santos-Martine G, Magana-Ortiz JE, Puente-Pinon SL (2013) Acute histoplasmosis in three Mexican sewer workers. Occup Med Lond 63:77–79
Sharma R, Sharma CL, Kapoor B (2005) Antibacterial resistance: current problems and possible solutions. Indian J Med Sci 59:120–129
Sherley M, Gardon DM, Collingnon PJ (2000) Variation in Antibiotics resistance profile in Enterobacteriaceae isolated from wild Australian mammels. Environ Microbiol 2:620–631
Thonda OA, Oluduro AO, Osuntokun OT, Aladejana OM, Ajadi FA (2018) Occurrence and multiple antibiotic susceptibility profile of bacterial pathogens isolated from patients chronic respiratory diseases. JMSCR 06(3):819–826. https://doi.org/10.18535/jmscr/v6i3.138
Zhao GM, Black S, Shinefield H (2003) Serotype distribution and antimicrobial resistance patterns in Streptococcus pneumoniae isolated from hospital pediatric patients with respiratory tract infections in Shanghai, China. Paediatr Infect Dis J 22:739–742
Author Information
Department of Biological Sciences, Faculty of Natural Sciences, Redeemer’s University, Ede, Nigeria