Genetic diversity revealed via molecular analysis of moroccan and foreign plum (Prunus domestica; Prunus salicina) genotypes from an ex-situ collection
Research Articles | Published: 29 August, 2022
First Page: 816
Last Page: 824
Views: 3459
Keywords: Plum tree, ISSR markers, Ex-situ collection, Genetic diversity, Polymorphism
Abstract
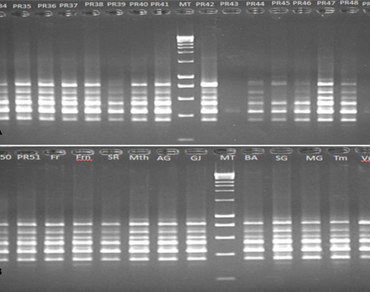
There is a need for plum breeding in Morocco to release efficient and adapted varieties, which requires the existence of genetic variability in the studied material. Molecular characterization of 29 plum cultivars, from a Moroccan ex-situ collection, was carried out using 20 Inter Simple Sequence Repeats (ISSR) primers. A high level of genetic variability was obtained among these plum cultivars with a polymorphism rate of 95.68% by finding 224 polymorphic bands among the 236 bands revealed by the primers used. Polymorphism information content (PIC), ranged from a minimum 0.19 to a maximum 0.47, showing that used ISSR markers are informative and relevant for discriminating the cultivars evaluated. Moreover, the resolving power (Rp) ranged from 0.72 to 1.70 with an average value of 1.17. The effective multiple ratio (EMR) ranged from 3.60 to 22 and the marker index (MI) ranged from 0.70 to 9.46. The similarity coefficient of ISSR data ranged from 0.136 to 0.619, with an average of 0.365. The ‘Stanley’ and ‘Prune d’ente’ varieties which are suitable for drying and belonging to the same species P. domestica are the closest genetically and the ‘INRA-PR43’ and ‘INRA-PR41’ genotypes are the most distinct. The results obtained showed that the ex-situ collection of plum is characterized by a high genetic diversity. Cluster analysis data revealed three main clusters and which are subdivided into 6 subgroups and a few isolated branches which presumably correspond to the different gene pools. The results confirm that ISSR detected high polymorphism level and represented high genetic variation in the plum ex-situ collection.
References
Ait bella Y, Bouda S, Haddioui A (2018) Phenotypic Diversity Analysis of Plum (Prunus domestica L.) Cultivars. Phytomorphology 68(34):93–101
Ait Bella Y, Bouda S, Khachtib Y, Haddioui A (2021) Genetic variability of cultivated plum (‘Prunus domestica L. and Prunus salicina’Lindl.) in Morocco assessed by ISSR markers. Aust J Crop Sci 15(6):948–954
Anderson JA, Churchill GA, Autroque JE, Tanksley SD, Swells ME (1993) Optimising selection for plant linkage map. Genome 36:181–186
Athanasiadis I, Nikoloudakis N, Hagidimitriou M (2013) Genetic relatedness among cultivars of the Greek plum Germplasm. Not Bot Horti Agrobo 41:49–498
Brzustowski J, Canada (2002) http://www.biology.ualberta.ca/jbrzusto/cluster.php. Accessed on 16 February, 2015
Cao W, Scoles G, Hucl P, Chibbar RN (2000) Phylogenetic relationships of five morphological groups of hexaploid wheat (Triticum aestivum L. em Thell.) based on RAPD analysis. Genome 43(4):724–727
Carrasco B, Díaz C, Moya M, Gebauer M, Gonzáles RG (2012) Genetic characterization of Japanese plum cultivars (Prunus salicina) using SSR and ISSR molecular markers. Ciencia e investigación agraria: revista latinoamericana de ciencias de la agricultura 39(3):533–543
Chao CT, Devanand PS, Chen J (2005) AFLP analysis of genetic relationships among Calathea species and cultivars. Plant Sci 168:1459–1469
Chesnokov YV, Artemyeva AM (2015) Evaluation of the measure of polymorphism information of genetic diversity. Сельскохозяйственная биология, (5 (eng))
De Vienne D (1998) Biotechnologies Végétales, p: 118. Les marqueurs moléculaires et leurs applications. CNED, Paris, France
Fanning K, Topp B, Russell D, Stanley R, Netzel M (2014) Japanese plums (Prunus salicina Lindl.) and phytochemicals-breeding, horticultural practices, postharvest storage, processing and bioactivity. J Sci Food Agric 94:2137–2147
FAO (2020) FAO Statistical Database, http://www.fao.org/faostat/en/#data/QC
Goulao L, Monte-Corvo L, Oliveira CM (2001) Phenetic characterization of plum cultivars by high multiplex ratio markers: Amplified fragment length polymorphisms and inter-simple sequence repeats. J Am Soc Hort Sci 126:72–77
Guerra ME, Rodrigo J (2015) Japanese plum pollination: A review. Sci Hortic 197:674–686
Hamdani A, Charafi J, Bouda S, Hssaini L, Adiba A, Razouk R (2021) Screening for water stress tolerance in eleven plum (Prunus salicina L.) Cultivars using agronomic and physiological traits. Sci Hortic 281:109992
Ilgin M, Kafkas S, Ercisli S (2009) Molecular characterization of plum cultivars by AFLP markers. Biotechnol Biotech Eq. 23:1189–1193
Kadir UY, Sezai E, Bayram MA, Yıldız D, Salih K (2009) Genetic Relatedness in Prunus Genus Revealed by Inter-simple Sequence Repeat Markers. 44:293–2972
Kumar D, Srivastava KK, Singh SR (2018) Morphological and horticultural diversity of plum varieties evaluated under Kashmir conditions. Trop Plant Res 5(1):77–82
Madre JF(2013) Logiciel mesurim Pro. Ver. 3,4. Académie d’Améins
Manco R, Basile B, Capuozzo C, Scognamiglio P, Forlani M, Rao R, Corrado G (2019) Molecular and Phenotypic Diversity of Traditional European Plum (Prunus domestica L.) Germplasm of Southern Italy. Sustainability 11(15):4112
Martins M, Tenreiro R, Oliveira MM (2003) Genetic relatedness of Portuguese almond cultivars assessed by RAPD and ISSR markers. Plant Cell Rep 22(1):71–78
Li M, Zhao Z, Miao XJ(2013) Genetic variability of wild apricot (Prunus armeniaca L.) populations in the Ili Valley as revealed by ISSR markers, https://doi.org/10.1007/s10722-013-9996-x
Liu W, Liu D, Zhang A, Feng C, Yang J, Yoon J, Li S (2007) Genetic Diversity and Phylogenetic Relationships among Plum Germplasm Resources in China Assessed with Inter-simple. Seq Repeat Markers 132(5):619–628
Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE(2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc. Natl. Acad Sci. USA, 99: 5836–5841
Okie WR, Ramming DW (1999) Plum breeding worldwide. HortTechnology 9:162–176
Ortiz A, Renaud R, Calzada I, Ritter E (1997) Analysis of plum cultivars with RAPD markers. J Hortic Sci 72:1–9
Sun Qi, Liao K, Geng W, Liu J, Nasir M, Liu H, Jia Y, Cao Q (2015) Analysis of Genetic Relationship in Prunus domestica L. Xinjiang Using ISSR Markers 16(3):447–453503
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98(1):107–112
Rohlf FJ (2002) NTSYS-pc Numerical Taxonomy System, ver. 2.1 Exeter. Publishing Ltd. Setauket, New York, USA
Roldan-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW(1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad Sci. USA, 81: 8014–8018
Salunkhe DK, Kadam SS (1995) Handbook of Fruit Science and Technology: Production, Composition, Storage, and Processing. CRC Press, Boca Raton, FL, USA. ISBN-13 978-0824796433
Stacewicz-Sapuntzakis M, Bowen PE, Hussain EA, Damayanti-Wood BI, Farnsworth NR (2001) Chemical composition and potential health effects of prunes: A functional food? Crit Rev Food Sci 41:251–286
Topp BL, Russell DM, Neumuller M, Dalbo MA, Liu W(2012) Fruit Breeding, Handbook of Plant Breeding; Badenes, M.L., Byrne, D.H., Eds.; Springer Science + Business Media: New York, NY, USA, ISBN 978-1-4419-0762-2
UPOV (1982) Guidelines for the conduct of tests for distinctness, homogeneity and stability. (Prunus salicina Lindl. and other diploid plums). Ginebra, Switzerland. 24 pp
Walali LDM, Skiredj A (2003) Apricot, plum, pear and apple trees. Transfer of technology in agriculture, Monthly information and liaison bulletin of PNTTA. Morocco, p 107
Woldring H (2000) On the origin of plums: a study of sloe, damson, cherry plum, domestic plums and their intermedi¬ates. Palaeohistoria 39(40):535–562
Wu W, Chen F, Yeh K, Chen J (2019) ISSR analysis of genetic diversity and structure of plum varieties cultivated in southern China. Biology 8(1):2
Author Information
National Agricultural Research Institute, Meknes, Morocco