Exploring the in silico studies of the endophyte fungus Phoma herbarum against mur enzymes of Staphylococcus aureus – a computational approach
Research Articles | Published: 23 October, 2023
First Page: 2470
Last Page: 2480
Views: 3890
Keywords: Endophytic fungi, Bioactive chemicals, Mur enzymes, Molecular docking, Drug-likeness
Abstract
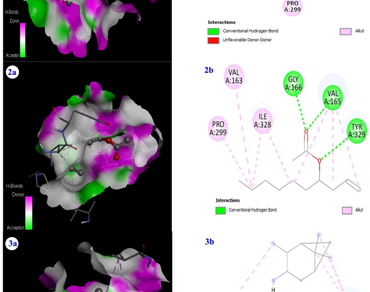
Endophyte fungus Phoma herbarum (NFCCI 4035) was previously isolated and identified from healthy leaf tissue of Meriandra bengalensis Benth plant from Manipur. We recently established the antibacterial activity of ethyl acetate extracts of endophyte fungus P. herbarum in vitro and identified some potentially active components using GC-MS. The current study aims to perform molecular docking of the discovered P. herbarum compounds against Mur enzymes (Mur A, Mur C, Mur D, and Mur F) of Staphylococcus aureus using Auto Dock Tools 1.5.6. SwissADME online tool was used to calculate the drug-likeness characteristics. According to the findings, only 14 of 23 bioactive compounds evaluated from P. herbarum extract adhered to Lipinski’s Rule of Five (LRo5) and had binding affinity ranged between − 3.5 to -6.5 (kcal/mol) with at least one of the four targeted Mur enzymes. When compared to all other bioactive compounds that interact with target Mur enzymes, 2-Palmitoylglycerol molecule demonstrated highest binding affinity (-6.5 kcal/mol) with Mur F ligase. However, (1R,2R,3R,5 S)-2,6,6-Trimethylbicyclo[3.1.1]heptan-3-amine has shown excellent binding affinity for all selected target (Mur A, Mur C, Mur D, and Mur F) enzymes. Also, this study confirmed that the bioactive compounds can be developed into more potent inhibitors targeting S. aureus-related infections.
References
Adetutu A, Olaniyi TD, Owoade OA (2021) GC-MS analysis and in silico assessment of constituents of Psidium guajava leaf extract against DNA gyrase of Salmonella enterica serovar Typhi. Inf Med Unlocked 26:100722. https://doi.org/10.1016/j.imu.2021.100722
Adnan M, Nazim Uddin Chy M, Mostafa Kamal AT, Azad MO, Paul A, Uddin SB, Barlow JW, Faruque MO, Park CH, Cho DH (2019) Investigation of the biological activities and characterization of bioactive constituents of Ophiorrhiza rugosa var. prostrata (D. Don) & Mondal leaves through in vivo, in vitro, and in silico approaches. Molecules. 24(7):1367
Chakravarthi V, Karthikeyan M, Lakshmanan L, Murugesan S, Arivuchelvan A, Sukumar K, Arulmozhi A, Jagadeeswaran A (2023) Computational study of Piper betle L. phytocompounds by in silico and ADMET analysis for prediction of potential xanthine oxidase inhibitory activity. BioRxiv. https://doi.org/10.1101/2023.01.13.523909
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Devi WS, Surendirakumar K, Singh MS (2022) Distribution of endophytic fungi associated with Meriandra bengalensis Benth. And assessment of their bioactive potential in vitro. Vegetos 35:995–1006
Engel H, Gutiérrez-Fernández J, Flückiger C, Martínez-Ripoll M, Mühlemann K, Hermoso JA, Hilty M, Hathaway LJ (2013) Heteroresistance to fosfomycin is predominant in Streptococcus pneumoniae and depends on the murA1 gene. Antimicrob Agents Chemother 57(6):2801–2808. https://doi.org/10.1128/AAC.00223-13
Freiberg C, Pohlmann J, Nell PG, Endermann R, Schuhmacher J, Newton B, Otteneder M, Lampe T, Häbich D, Ziegelbauer K (2006) Novel bacterial acetyl coenzyme a carboxylase inhibitors with antibiotic efficacy in vivo. Antimicrob Agents Chemother 50(8):2707–2712
Frisvad JC, Houbraken J, Popma S, Samson RA (2013) Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiol Lett 339:77–92
Funes Chabán M, Hrast M, Frlan R, Graikioti DG, Athanassopoulos CM, Carpinella MC (2021) Inhibition of Mur a enzyme from Escherichia coli and Staphylococcus aureus by Diterpenes from Lepechinia Meyenii and their synthetic analogs. Antibiotics 10(12):1535. https://doi.org/10.3390/antibiotics10121535
Gaur V, Bera S (2022) Recent developments on UDP-N-acetylmuramoyl-L-alanine-D-gutamate ligase (mur D) enzyme for antimicrobial drug development: an emphasis on in-silico approaches. Curr Res Pharmacol Drug Disc 3:100137. https://doi.org/10.1016/j.crphar.2022.100137
Hashmi AA, Naz S, Hashmi SK, Irfan M, Hussain ZF, Khan EY, Asif H, Faridi N (2019) Epidermal growth factor receptor (EGFR) overexpression in triple-negative breast cancer: association with clinicopathologic features and prognostic parameters. Surg Exp Pathol 2(6):1–7
Hrast M, Sosič I, Šink R, Gobec S (2014) Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem 55:2–15
Jha RK, Khan RJ, Amera GM, Singh E, Pathak A, Jain M, Muthukumaran J, Singh AK (2020) Identification of promising molecules against MurD ligase from Acinetobacter baumannii: insights from comparative protein modelling, virtual screening, molecular dynamics simulations and MM/PBSA analysis. J Mol Model 26:1–7
Kotnik M, Humljan J, Contreras-Martel C, Oblak M, Kristan K, Hervé M, Blanot D, Urleb U, Gobec S, Dessen A, Solmajer T (2007) Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase. J Mol Biol 370(1):107–115
Kouidmi I, Levesque RC, Paradis-Bleau C (2014) The biology of Mur ligases as an antibacterial target. Mol Microbiol 94(2):242–253
Laddomada F, Miyachiro MM, Dessen A (2016) Structural insights into protein-protein interactions involved in bacterial cell wall biogenesis. Antibiotics 5(2):14. https://doi.org/10.3390/antibiotics5020014
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Liu Y, Breukink E (2016) The membrane steps of bacterial cell wall synthesis as antibiotic targets. Antibiotics 5(3):28. https://doi.org/10.3390/antibiotics5030028
Mehjabeen H, Dil USC, Jacy Farhana, Mohammed TA, Ananya C, Shamima I, Adnan M (2013) Identification of potential targets in Staphylococcus aureus N315 using computer aided protein data analysis. Bioinformation 9(4):187–192
Mihalovits LM, Ferenczy GG, Keseru GM (2019) Catalytic mechanism and covalent inhibition of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA): implications to the design of novel antibacterials. J Chem Inf Model 59:5161–5173
Mol CD, Brooun A, Dougan DR, Hilgers MT, Tari LW, Wijnands RA, Knuth MW, McRee DE, Swanson RV (2003) Crystal structures of active fully assembled substrate-and product-bound complexes of UDP-N-acetylmuramic acid: L-alanine ligase (MurC) from Haemophilus influenzae. J Bacteriol 185(14):4152–4162
Mousa WK, Raizada MN (2013) The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front Microbiol 4:65. https://doi.org/10.3389/fmicb.2013.00065
Ononamadu CJ, Ibrahim A (2021) Molecular docking and prediction of ADME/drug-likeness properties of potentially active antidiabetic compounds isolated from aqueous-methanol extracts of Gymnema sylvestre and Combretum micranthum. Biotechnologia 102(1):85–99
Rani N, Kumar C, Arunachalam A, Lakshmi PTV (2018) Rutin as a potential inhibitor to target peptidoglycan pathway of Staphylococcus aureus cell wall synthesis. Clin Microbiol Infect Dis 3:15761. https://doi.org/10.15761/CMID.1000142
Rath S, Jagadeb M, Bhuyan R (2021) Molecular docking of bioactive compounds derived from Moringa oleifera with p53 protein in the apoptosis pathway of oral squamous cell carcinoma. Genomics Inf 19(4):e46. https://doi.org/10.5808/gi.21062
Srinivasan K, Altemimi AB, Narayanaswamy R, Vasantha Srinivasan P, Najm MA, Mahna N (2023) GC-MS, alpha‐amylase, and alpha‐glucosidase inhibition and molecular docking analysis of selected phytoconstituents of small wild date palm fruit (Phoenix pusilla). Food Sci Nutr 1–14. https://doi.org/10.1002/fsn3.3489
Staniek A, Woerdenbag J, Kayser O (2008) Endophytes exploiting biodiversity for the improvement of natural product-based drug discovery. J Plant Interact 3:75–98
Suresh E, Sureshkumar P (2019) Antimicrofouling activity of Calotropis gigantea (L). R. Br. Indian J Geo-Mar Sci 48(12):1843–1848
Tiwari P, Srivastava Y, Bae H (2021) Endophytes: Trend of pharmaceutical design of endophytes as anti-infective. Curr Top Med Chem 21:1572–1586
Umamaheswari A, Pradhan D, Hemanthkumar M (2010) Virtual screening for potential inhibitors of homology modeled Leptospira interrogans MurD ligase. J Chem Biol 3:175–187
Vollmer W, Blanot D, De Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167
Yashodeep S, Iqrar Ahmad, Sanjay S, Harun P (2021) The mur enzymes chink in the armour of Mycobacterium tuberculosis cell wall. Eur J Med Chem 222(23–24):113568
Author Information
Department of Life Sciences (Botany), Manipur University, Canchipur, Manipur, India