Evaluation of genetic diversity using biochemical markers in sugarcane germplasm collection
Research Articles | Published: 28 August, 2023
First Page: 1543
Last Page: 1551
Views: 3147
Keywords: Sugarcane germplasm collection, Protein profiling, Biochemical markers, Genetic diversity, n Saccharum species, SDS-PAGE analysis, Phylogenetic relationship
Abstract
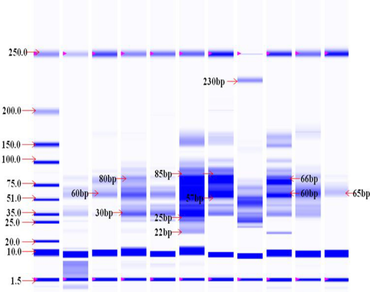
Sugarcane (Saccharum spp.) is a genetically complex crop of great economic importance in tropical and subtropical countries around the world. Currently, main concern is to characterize sugarcane germplasm based on protein profiling which provide key information on genetic diversity that can be used in breeding for yield and quality traits. The present study aimed to evaluate complement of leaf proteins using SDS-PAGE technique in sugarcane accessions as an alternative biochemical marker system for DNA-based markers used in genetic diversity analysis. DNA-based molecular markers are usually employed in genetic diversity analysis, but assays based on these markers are relatively expensive, time-consuming and tedious operations than alternative protein-based biochemical markers. To conduct experiment, the protein extract was subjected to SDS–polyacrylamide gel electrophoresis using a rapid and economical Experion automated electrophoretic resolution system. The germplasm accessions represented different origins in the Indian subcontinent and four clones from Jawa, Indonesia. A total of 748 protein bands were obtained, of which 452 (54.0%) bands were found polymorphic in the entire set of 50 accessions used in the study. The dendrogram constructed using UPGMA algorithm divided all the accessions into two major groups consisting five clusters. The clustering pattern indicated that genetic diversity among Saccharum spontaneum and subtropical accessions was greater, which could be due to their unique genetic constitution. Genetic diversity between S. spontaneum and UPCSR or S. officinarum or S. robustum accessions was less that indicates shared protein pool, which may be due to common ancestry and exchange of germplasm among different breeding groups. Analysis of the dendrogram was not allowed to distinguish interspecific hybrids from foreign commercial hybrids originated in India and Indonesia. Accessions from extensive geographic sources differed considerably, but it was difficult to establish any relationship between the place of origin and the nature of clustering. The obtained protein profiling data indicated a biochemical polymorphism and segregating clustering pattern that may be informative for phylogenetic studies. The degree of genetic diversity assessed using biochemical/protein markers will be useful in future sugarcane breeding programs to select promising parental lines.
References
Alipour H, Rezai A, Meibodi SAM, Taheri M (2002) Evalution of genetic variation in soybean lines using seed protein electrophoresis. J Sci Technol Agric Nat Resour 5(4):85–96
Bailey DC (1983) Isozyme variation and plant breeders ‘rights. In: Tanksely SD, Orton TJ (eds) Isozymes in plant genetics and breeding, part A. Elsevier, Amsterdam, pp 425–440
Bajpai A, Srivastva S (1999) SDS protein markers as an aid to differentiate sugarcane clones. In: National Seminar on Advance in sugarcane technology. Nov 2–3, 1999. I.I.S.R., Lucknow, p 52
Barnabas L, Ramadass A, Amalraj RS, Palaniyandi M, Rasappa V (2015) Sugarcane proteomics: an update on current status, challenges, and future prospects. Proteomics 15(10):1658–1670
Beckstrom-Sternberg SM (1989) Two-dimensional gel electrophoresis as an taxonomic tool: evidence from the centrospermeae. Biochem Syst Ecol 17:573–582
Celis JE, Bravo R (1984) Two dimensional gel electrophoresis of proteins; methods and applications. Academic Press, New York
Champs G, Fde-J V, Augustin E, Irigon D (1994) Morphological and electrophoretical chacterization of twenty soybean cultivars. Pesqui Agropecu Bras 29(11):1779–1787
Chandra P, Singh RK, Singh IS, Singh SB (2001) Possibility of identification and characterization of sugarcane cultivars on the basis of protein profile. Sugar Tech 3:59–62
Das S, Mukherjee KK (1995) Comparative study on seed proteins of Ipomea. Seed Sci Technol 23:501–509
Eksomtramage F, Paulet JL, Noyer P, Feldmann P, Glaszmann JC (1992) Utility of isozymes in sugarcane breeding. Sugarcane 3:14–21
Gantait S, Mandal N, Das PK (2009) Diversity of blackgram (Vigna mungo L. Hepper) genotypes assessed through morphological and biochemical approaches. J Trop Agric 47(1):80–83
Gepts P (1989) Genetic diversity of seed storage proteins in plants. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetics resources. Sinauer Associates Inc., Sunderland, pp 64–82
Ghafoor A, Gulbaaz FN, Afzal M, Ashraf M, Arshad M (2003) Inter-relationship between SDS-PAGE markers and agronomic traits in chickpea (Cicer arietinum). Pak J Bot 35(4):613–624
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487
Hemaprabha G, Rangasamy SR (2001) Genetic similarity among five species of Saccharum based on isozyme and RAPD markers. Indian J Genet 61:341–347
Ismail A, Mosa KA, Ali MA, Helmy M (2020) Biochemical and molecular markers: unraveling their potential role in screening germplasm for thermotolerance. In: Heat stress tolerance in plants: physiological, molecular and genetic perspectives. Wiley, pp 47–76. https://doi.org/10.1002/9781119432401. ISBN: 9781119432401
Javaid A, Ghafoor A, Anwar R (2004) Seed storage protein electrophoresis in groundnut for evaluating genetic diversity. Pak J Bot 36:25–29
Jha SS, Ohri D (1996) Phylogenetic relationship of Cajanus cajan (L.) Millsp. (Pigeonpea) and its wild relatives based on seed protein profiles. Genet Resour Crop Evol 43:275–281
Jiang S, Liu S, Wu ZC, C, (2016) Developing protocols of Tricine-SDS-PAGE for separation of polypeptides in the mass range 1–30 kDa with minigel electrophoresis system. Int J Electrochem Sci 11:640–649
Joshi R, Shukla A, Kumar P (2013) In vitro water deficit stress induced genotypic alterations in protein profile among aromatic rice varieties. Ann Plant Sci 02(10):455–458
Khurshid M (2022) Assessment of genetic diversity of sugarcane (Saccharum spp.) genotypes through biochemical approach. Agric Sci Dig A Res J 42(1):38–42
Kohila S, Gomathi R (2018) Adaptive physiological and biochemical response of sugarcane genotypes to high-temperature stress. Indian J Plant Physiol 23(2):245–260
Lowry OH, Rosebrough NJ, Farr AL (1951) Protein measurement with folin phenol reagent, Lowry protein. J Biol Chem 193:165–275
Malik MFA, Afsari S, Qureshi AM, Khan MR, Javed A (2009) Evolution of genetic diversity in Soyabean (Glycine max) lines using seed protein electrophoresis. Aust J Crop Sci 3(2):107–112
Medar RA, Rajpurohit VS, Ambekar AM (2019) Sugarcane crop yield forecasting model using supervised machine learning. Int J Intell Syst Appl 11(8):11
Mehdi F, Kazim ALİ, Nesheman HUMA, Hussain I, Azhar A, Galani S (2020) Comparative biochemical analysis of high and low sucrose accumulating sugarcane varieties at formative stage under heat stress. J Agric Sci 26:78–86
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. J Comput Appl Biosci 12:357–358
Rao VP, Sengar RS, Singh RB (2021) Identification of salt tolerant sugarcane cultivars through phenotypic, physiological and biochemical studies under abiotic stress. Plant Physiol Rep 26(2):256–283
Saxena P, Srivastava RP, Sharma ML (2010) Studies on salinity stress tolerance in sugarcane varieties. Sugar Tech 12:59–63
Singh RS, Jain SK, Qualset CO (1973) Protein electrophoresis as an aid to oat variety identification. Euphytica 22(1):98–105
Singh RK, Chandra P, Singh DN, Singh RG, Singh SB (1999) Morphologically distinct mutants in sugarcane and their protein profiles. Sugar Tech 3:93–97
Singh RK, Singh RB, Singh SP, Sharma ML (2011) Identification of sugarcane microsatellites associated to sugar content in sugarcane and transferability to other cereal genomes. Euphytica 182(3):335–354
Singh RK, Singh RB, Singh SP, Mishra N, Rastogi J, Sharma ML, Kumar A (2013) Genetic diversity among Saccharum spontaneum clones and commercial hybrids through SSR markers. Sugar Tech 15(2):109–115
Singh RB, Singh B, Singh RK (2017) Study of genetic diversity of sugarcane (Saccharum) species and commercial varieties through TRAP molecular markers. Indian J Plant Physiol 22:332–338
Singh RB, Singh B, Singh RK (2018) Evaluation of genetic diversity in Saccharum species clones and commercial varieties employing molecular (SSR) and physiological markers. Indian J Plant Genet Resour 31(1):17–26
Singh RB, Singh B, Singh RK (2019) Development of potential dbEST-derived microsatellite markers for genetic evaluation of sugarcane and related cereal grasses. Ind Crop Prod 128:38–47
Singh RB, Mahenderakar MD, Jugran AK, Singh RK, Srivastava RK (2020) Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene 753:144800
Siva R, Kumar K, Rajasekaran C (2013) Genetic diversity study of important Indian rice genotypes using biochemical and molecular markers. Afr J Biotech 12(10):1004–1009
Srivastava S, Gupta PS (2002) SDS and Native page protein profile for identification and characterization of elite sugarcane genotypes. Sugar Tech 4:143–147
Vanlalsanga Y, Tunginba S (2019) Genetic diversity and population structure in upland rice (Oryza sativa L.) of Mizoram, north East India as revealed by morphological, biochemical and molecular markers. Biochem Genet 57:421–442
Ward JH Jr (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58(301):236–244
Author Information
Department of Biotechnology, School of Applied Sciences, REVA University, Bangalore, India