Efficient decolourization of toluidine blue and methylene blue using Bacillus cereus
Research Articles | Published: 18 October, 2023
First Page: 2439
Last Page: 2447
Views: 3436
Keywords: Methylene blue, n Bacillus cereusn , Fourier Transform Infrared Spectroscopy, Dye decolorization
Abstract
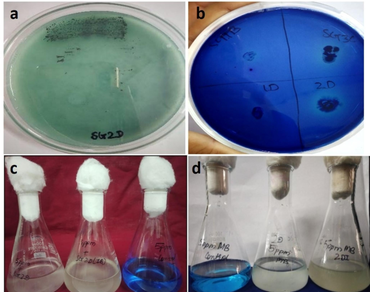
Dyes released from different industries have a negative impact on both human health and the environment. The use of microbial cultures for the biodegradation and decolorization of synthetic dyes is a cutting-edge, rapid, cost-effective, and eco-friendly approach. The present study was conducted to examine the decolorization of two synthetic basic dyes (toluidine blue and methylene blue) using an indigenous bacterium, Bacillus cereus. Quantification of dye decolorization was done using free and immobilized bacterial culture in a minimal salt medium augmented with different concentrations of dyes (5, 10, 15, and 20 ppm) after 3 and 5 days of incubation. The maximum dye removal efficiency of the free-living and immobilized culture of Bacillus cereus for methylene and toluidine blue were 94.92 and 91.18 and 94.92, and 93.62%, respectively after 5 days of incubation. The optimum concentrations of methylene and toluidine blue tolerated by the free and immobilized cultures of Bacillus cereus were 5 and 15 ppm and 15 and 20 ppm, respectively. Immobilized Bacillus cereus was more effective in dye decolorization as compared to a free bacterial culture. Significant induction in laccase activity was also observed by the test bacteria. The appearance and disappearance of peaks in the treated dye samples and the stretching of bonds at various wave numbers as compared to the control were correlated with the dye decolorization efficiency of Bacillus cereus.
References
Afrin S, Shuvo, Banjir HR, Sultana, Islam F, Rus’d AA, Begum S, Hossain MN (2021) The degradation of textile industry dyes using the effective bacterial consortium. Heliyon 7(10). https://doi.org/10.1016/j.heliyon.2021.e08102
Akansha K, Chakraborty D, Sachan SG (2019) Decolorization and degradation of methyl orange by Bacillus stratosphericus SCA1007. Biocat and Agricultural Biotechnol 18:101044. https://doi.org/10.1016/j.bcab.2019.101044
Alaya V, Kodi RK, Ninganna E, Gowda B, Shivanna MB (2021) Decolorization of Malachite green dye by Stenotrophomonas maltophilia a compost bacterium. Bull Natl Res Cent 45:81. https://doi.org/10.1186/s42269-021-00518-w
Alhassani HA, Rauf MA, Ashraf SS (2007) Efficient microbial degradation of Toluidine Blue dye by Brevibacillus sp. Dyes Pigm 75(2):395–400
Arora DS, Sandhu DK (1985) Laccase production and wood degradation by a white-rot fungus Daedalea flavida. Enzyme and Microbial Technol 7(8):405–408. https://doi.org/10.1016/0141-0229(85)90131-0
Babu SS, Mohandass C, Raj AV, Rajasabapathy R, Dhale MA (2013) Multiple approaches towards decolorization and reuse of a textile dye (VB-B) by a marine bacterium Shewanella decorations. Water Air Soil Pollut 224(4):1–12
Bharagava RN, Mani S, Mulla SI, Saratale GD (2018) Degradation and decolourization potential of an ligninolytic enzyme producing Aeromonas hydrophila for crystal violet dye and its phytotoxicity evaluation. Ecotoxicol and Environmental Saf 156:166–175. https://doi.org/10.1016/j.ecoenv.2018.03.012
Bhatt P, Pandey SC, Joshi S, Chaudhary P, Pathak VM, Huang Y, Wu X, Zhou Z, Chen S (2022) Nanobioremediation: a sustainable approach for the removal of toxic pollutants from the environment. J Hazard Mater 427:128033. https://doi.org/10.1016/j.jhazmat.2021.128033
Bilinska L, Blus K, Gmurek M, Ledakowicz S (2019) Coupling of electrocoagulation and ozone treatment for textile wastewater reuse. Chem Eng J 358:992–1001. https://doi.org/10.1016/j.cej.2018.10.093
Blanquez A, Rodríguez J, Brissos V, Mendes S, Martins LO, Ball AS, Arias ME, Hernández M (2019) Decolorization and detoxification of textile dyes using a versatile Streptomyces laccase-natural mediator system. Saudi J of Biological Sci 26(5). https://doi.org/10.1016/j.sjbs.2018.05.020
Das GS, Shim JP, Bhatnagar A, Tripathi KM, Kim T (2019) Biomass-derived carbon quantum dots for visible-light-induced photocatalysis and label-free detection of Fe (III) and ascorbic acid. Sci Rep 9(1):15084. https://doi.org/10.1038/s41598-019-49266-y
Ekambaram SP, Perumal SS, Annamalai U (2016) Decolorization and biodegradation of remazol reactive dyes by Clostridium species. 3 Biotech 6(1):20. https://doi.org/10.1007/s13205-015-0335-0
Fathy R, Omara AM (2023) Isolation and optimization of polyphosphate accumulating bacteria for bio-treatment of phosphate from industrial wastewater. Environ Technol 1–32. https://doi.org/10.1080/09593330.2023.2248558
Gangola S, Joshi S, Kumar S, Sharma B, Sharma A (2021) Differential proteomic analysis under pesticides stress and normal conditions in Bacillus cereus 2D. PLoS ONE 16(8):e0253106. https://doi.org/10.1371/journal.pone.0253106
Guembri M, Neifar M, Saidi M, Ferjani R, Chouchane H, Mosbah A, Ouzari HI (2021) Decolorization of textile azo dye Novacron Red using bacterial monoculture and consortium: response surface methodology optimization. Water Environ Res 93(8):1346–1360. https://doi.org/10.1002/wer.1521
Ghattavi S, Nezamzadeh-Ejhieh A (2020) GC-MASS detection of methyl orange degradation intermediates by AgBr/g-C3N4: experimental design, bandgap study, and characterization of the catalyst. Intern J of Hydrogen En 45:24636–24656. https://doi.org/10.1016/j.ijhydene.2020.06.207
Hanis KKA, Nasri AM, Farahiyah WW, Rabani MM (2020) Bacterial Degradation of Azo Dye Congo Red by Bacillus sp. In Journal of Physics: Conference Series (Vol. 1529, No. 2, p. 022048). IOP Publishing
Horitsu H, Takada M, Idaka E, Tomoyeda M, Ogawa T (1977) Degradation of p-aminoazobenzene by Bacillus subtilis. Eur J Appl Microbiol Biotechnol 4(3):217–224
Jayamohan H, Smith YR, Gale BK, Mohanty SK, Misra M (2016) Photocatalytic microfluidic reactors utilizing titania nanotubes on titanium mesh for degradation of organic and biological contaminants. J Environ Chem Eng 4(1):657–663. https://doi.org/10.1016/j.jece.2015.12.018
John J, Dineshram R, Hemalatha KR, Dhassiah MP, Gopal D, Kumar A (2020) Bio-decolorization of synthetic dyes by a Halophilic Bacterium Salinivibrio sp. Front Microbiol 11:594011. https://doi.org/10.3389/fmicb.2020.594011
Juybar M, Khorrami MK, Garmarudi AB, Zandbaaf S (2020) Determination of acidity in metal incorporated zeolites by infrared spectrometry using artificial neural network as chemometric approach. Spectrochim Acta Part A Mol Biomol Spectrosc 228:117539. https://doi.org/10.1016/j.saa.2019.117539
Kalyani DC, Patil PS, Jadhav JP, Govindwar SP (2008) Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresource Technol 99(11):4635–4641. https://doi.org/10.1016/j.biortech.2007.06.058
Karim ME, Dhar K, Hossain MT (2018) Decolorization of textile reactive dyes by bacterial monoculture and consortium screened from textile dyeing effluent. J of Genetic Engineering and Biotechnol 16(2):375–380. https://doi.org/10.1016/j.jgeb.2018.02.005
Khairun KAH, Muhammad ARN, Farahiyah WKW, Rabani MY (2019) Bacterial Degradation of Azo Dye Congo Red by Bacillus sp. Journal of Physics: Conference Series IOP Publishing 1529:022048. https://doi.org/10.1088/1742-6596/1529/2/022048
Kilany M (2017) Isolation, screening, and molecular identification of novel bacterial strain removing methylene blue from water solutions. Appl Water Sci 7(7):4091–4098. https://doi.org/10.1007/s13201-017-0565-x
Kim J, Chang S, Chung W (2022) Biodegradation of methylene blue using a novel lignin peroxidase enzyme producing bacteria, named Bacillus sp. React3, as a promising candidate for dye-contaminated wastewater treatment. Fermentation 8(5):190. https://doi.org/10.3390/fermentation8050190
Kishor R, Purchase D, Saratale GD, Saratale RG, Ferreira LFR, Bilal M, Bharagava RN (2021) Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. J Environ Chem Eng 9(2):105012. https://doi.org/10.1016/j.jece.2020.105012
Kumar A, Sharma A, Chaudhary P, Gangola S (2021) Chlorpyrifos degradation using binary fungal strains isolated from industrial waste soil. Biologia. https://doi.org/10.1007/s11756-021-00816-8
Kumar M, Kumari A, Vaghani BP, Chaudhary DR (2023) Dye degradation by early colonizing marine bacteria from the Arabian Sea, India. Arch Microbiol 205(4):160. https://doi.org/10.1007/s00203-023-03496-x
Kumar NM, Saravanan D (2015) Isolation of dye degrading bacteria from textile effluent. J of Chemical and Pharmaceutical Res 2214–2218
Lalnunhlimi S, Krishnaswamy V (2016) Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Brazilian J of Microbiol 47(1):39–46. https://doi.org/10.1016/j.bjm.2015.11.013
Liu Y, Shao Z, Reng X, Zhou J, Qin W (2021) Dye-decolorization of a newly isolated strain Bacillus amyloliquefaciens W36. World J of Microbiology and Biotechnol 37(1):1–11. https://doi.org/10.1007/s11274-020-02974-4
Mirsalari S, Nezamzadeh-Ejhieh A, Massah AA (2022) A designed experiment for CdS-AgBr photocatalyst toward methylene blue. Environ Sci Pollut Res 29:33013–33032. https://doi.org/10.1007/s11356-021-17569-1
Mirsalari SA, Nezamzadeh-Ejhieh A (2020) Focus on the photocatalytic pathway of the CdS-AgBr nano-catalyst by using the scavenging agents. Sep Purif Technol 250:117235. https://doi.org/10.1016/j.seppur.2020.117235
Mohanty SS, Kumar A (2021) Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci Rep 11:7678. https://doi.org/10.1038/s41598-021-87227-6
Moharrery L, Otadi M, Miraly N, Zangeneh RMM, Amiri R (2019) Degradation of toluidine red, an oil soluble azo dye by Halomonas strain IP8 at alkaline condition. Chem Eng Commun 206(1):61–68
Nezamzadeh-Ejhieh A, Karimi-Shamsabadi M (2014) Comparison of photocatalytic efficiency of supported CuO onto micro and nano particles of zeolite X in photodecolorization of Methylene blue and Methyl orange aqueous mixture. Appl Catal A 477:83–92. https://doi.org/10.1016/j.apcata.2014.02.031
Nho SW, Cui X, Kweon O et al (2021) Phylogenetically diverse bacteria isolated from tattoo inks, an azo dye-rich environment, decolorize a wide range of azo dyes. Ann Microbiol 71:35. https://doi.org/10.1186/s13213-021-01648-2
Olukanni OD, Adenopo A, Awotula AO, Osuntoki AA (2013) Biodegradation of malachite green by extracellular laccase producing Bacillus thuringiensis RUN1. J of Basic and Applied Sciences 9:543–549
Pham VHT, Kim J, Chang S, Chung W (2022) Biodegradation of Methylene Blue using a novel lignin peroxidase enzyme producing bacteria, named Bacillus sp. React3, as a promising candidate for dye-contaminated wastewater treatment. Fermentation 8(5):190. https://doi.org/10.3390/fermentation8050190
Phugare SS, Kalyani DC, Surwase SN, Jadhav JP (2011) Ecofriendly degradation, decolorization and detoxification of textile effluent by a developed bacterial consortium. Ecotoxicol and Environ Safety 74(5):1288–1296. https://doi.org/10.1016/j.ecoenv.2011.03.003
Senobari S, Nezamzadeh-Ejhieh A (2018a) A comprehensive study on the enhanced photocatalytic activity of CuO-NiO nanoparticles: designing the experiments. J of Mol Liquids 261:208–217. https://doi.org/10.1016/j.molliq.2018.04.028
Senobari S, Nezamzadeh-Ejhieh A (2018b) A pn junction NiO-CdS nanoparticles with enhanced photocatalytic activity: a response surface methodology study. J of Mol Liquids 257:173–183. https://doi.org/10.1016/j.molliq.2018.02.096
Sharma V, Upadhyay LS, Vasanth D (2020) Extracellular thermostable laccase-like enzymes from Bacillus licheniformis strains: production, purification and characterization. Appl Biochem Microbiol 56:420–432. https://doi.org/10.1134/S0003683820040146
Tahir U, Nawaz S, Khan UH, Yasmin A (2021) Assessment of biodecolorization potentials of biofilm forming bacteria from two different genera for mordant black 11 dye. Bioremediat J 25:252–270. https://doi.org/10.1080/10889868.2021.19119
Author Information
Department of Microbiology, Govind Ballabh Pant University of Agriculture & Technology, Pantnagar, India