Computational screening of natural compounds from Salvia plebeia R. Br. for inhibition of SARS-CoV-2 main protease
Zackria Afraa Aqeel, Pattabiraman Ramya, Murthy T. P. Krishna, Kumar S. Birendra, Mathew Blessy Baby, Biju Vinai George
Research Articles | Published: 19 October, 2021
First Page: 345
Last Page: 359
Views: 4050
Keywords: Main protease, Salvia plebeia R. Br., Rutin, Plebeiosides B, ADMET, PASS, Molecular docking, Molecular dynamic simulation
Abstract
The novel Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) has emerged to be the reason behind the COVID-19 pandemic. It was discovered in Wuhan, China and then began spreading around the world, impacting the health of millions. Efforts for treatment have been hampered as there are no antiviral drugs that are effective against this virus. In the present study, we have explored the phytochemical constituents of Salvia plebeia R. Br., in terms of its binding affinity by targeting COVID-19 main protease (Mpro) using computational analysis. Molecular docking analysis was performed using PyRx software. The ADMET and drug-likeness properties of the top 10 compounds showing binding affinity greater than or equal to − 8.0 kcal/mol were analysed using pkCSM and DruLiTo, respectively. Based on the docking studies, it was confirmed that Rutin and Plebeiosides B were the most potent inhibitors of the main protease of SARS-CoV-2 with the best binding affinities of − 9.1 kcal/mol and − 8.9 kcal/mol, respectively. Further, the two compounds were analysed by studying their biological activity using the PASS webserver. Molecular dynamics simulation analysis was performed for the selected protein–ligand complexes to confirm their stability at 300 ns. MM-PBSA provided the basis for analyzing the affinity of the phytochemicals towards Mpro by calculating the binding energy, and secondary structure analysis indicated the stability of protease structure when it is bound to Rutin and Plebeiosides B. Altogether, the study identifies Rutin and Plebeiosides B to be potent Mpro inhibitors of SARS-CoV-2.
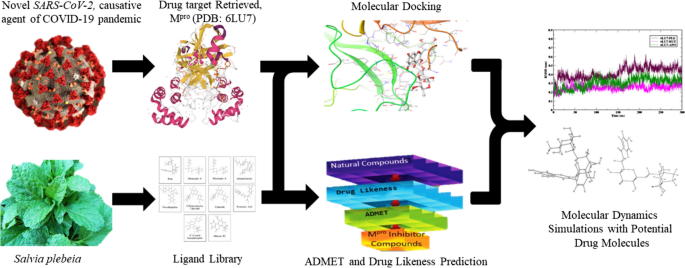
Graphic abstract

References
Amin SA, Jha T (2020) Fight against novel coronavirus: a perspective of medicinal chemists. Eur J Med Chem 201:112559
Araruna MKA et al (2012) Evaluation of antibiotic & antibiotic modifying activity of pilocarpine & rutin. Indian J Med Res 135(2):252
Bang S et al (2016) Antiviral activities of compounds from aerial parts of Salvia plebeia R. Br. J Ethnopharmacol 192:398–405
Bang S et al (2018) Anti-influenza effect of the major flavonoids from Salvia plebeia R.Br. via inhibition of influenza H1N1 virus neuraminidase. Nat Prod Res 32(10):1224–1228
Das S, Koner BC (2020) Pre-analytical, analytical, and post-analytical considerations while processing samples of COVID-19 patients: perspective from a clinical chemistry laboratory in India. Asian J Med Sci 11(5):112–115
Das P, Majumder R, Mandal M, Basak P (2020) In-silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of calendula officinalis. J Biomol Struct Dyn 39:1–16
Fauci AS, Clifford Lane H, Redfield RR (2020) Covid-19—navigating the uncharted. N Engl J Med 382(13):1268–1269
Fereidoonnezhad M et al (2018) Multitarget drug design, molecular docking and PLIF studies of novel tacrine−coumarin hybrids for the treatment of Alzheimer’s disease. Iran J Pharm Res 17(4):1217–1228
Ghasemnezhad A, Ghorbanzadeh A, Sarmast MK, Ghorbanpour M (2020) A review on botanical, phytochemical, and pharmacological characteristics of Iranian Junipers (Juniperus Spp.). Plant-derived bioactives: production, properties and therapeutic applications. Springer, Singapore
Gibaldi M, Levy G (1976) Pharmacokinetics in clinical practice: I. Concepts. JAMA J Am Med Assoc 235(17):1864–1867
Gil C et al (2020) COVID-19: drug targets and potential treatments. J Med Chem. https://doi.org/10.1021/acs.jmedchem.0c00606
Gupta S et al (2020) Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1776157
Han Y et al (2019) In silico ADME and toxicity prediction of ceftazidime and its impurities. Front Pharmacol 10(APR):434
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discovery 14(2):111–129
Ibrahim MAA, Abdeljawaad KAA, Abdelrahman AHM, Hegazy MEF (2020) Natural-like products as potential SARS-CoV-2 Mpro inhibitors: in-silico drug discovery. J Biomol Struct Dyn 39:1–13
Jacob RB, Andersen T, Mcdougal OM (2012) “Accessible high-throughput virtual screening molecular docking software for students and educators” ed Fran Lewitter. PLoS Comput Biol 8(5):e1002499
Jin Z et al (2020) Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582(7811):289–293. https://doi.org/10.1038/s41586-020-2223-y
JP, Lin, et al (2009) Rutin inhibits the proliferation of murine leukemia WEHI-3 cells in vivo and promotes immune response in vivo. Leuk Res 33(6):823–828
Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M (2020) From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol 92(6):660–666
Karami M, Jalali C, Mirzaie S (2017) Combined virtual screening, MMPBSA, molecular docking and dynamics studies against deadly anthrax: an in silico effort to inhibit bacillus anthracis nucleoside hydrolase. J Theor Biol 420:180–189
Krishna S, Kumar SB, Krishna Murthy TP, Murahari M (2021) Structure-based design approach of potential BCL-2 inhibitors for cancer chemotherapy. Comput Biol Med 134:104455
Kumar S, Pandey AK (2013) Chemistry and biological activities of flavonoids: an overview. Sci World J 2013:1–16
Kumar SB et al (2021) Screening of natural compounds from Cyperus Rotundus Linn against SARS-CoV-2 main protease (Mpro): an integrated computational approach. Comput Biol Med 134:104524–104524
Kumar B et al (2021) In silico screening of therapeutic potentials from strychnos nux-vomica against the dimeric main protease (Mpro) structure of SARS-CoV-2. J Biolmol Struct Dyn. https://doi.org/10.1080/07391102.2021.1902394
Kumari R, Kumar R, Lynn A (2014) G-Mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962. https://doi.org/10.1021/ci500020m
Li H et al (2020) Coronavirus Disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrobial Agents 55(5):105951
Liang Y et al (2020) Salvia plebeia R. Br.: an overview about its traditional uses, chemical constituents, pharmacology and modern applications. Biomed Pharmacother 121(219):109589
Lindahl, Abraham, Hess, van der Spoel (2019) GROMACS 2019.4 manual
Ma Q et al (2014) Chemistry and Pharmacology of Salvia Plebeia R. Brown (Lamiaceae ). J Chem Pharm Res 6(10):777–783
Mansoor A, Mahabadi N (2021) Volume of Distribution. In: StatPearls [Internet]. StatPearls Publishing, Treasure Island (FL). https://www.ncbi.nlm.nih.gov/books/NBK545280/
Marathe SA, Datey AA (2012) Herbal cocktail as anti-infective: promising therapeutic for the treatment of viral diseases. Recent Pat Anti-Infective Drug Discov 7(2):123–132
Mason RJ (2020) Pathogenesis of COVID-19 from a cell biology perspective. Eur Respir J 55(4):2000607
Murugesan S, Venkateswaran MR, Jayabal S, Periyasamy S (2020) Evaluation of the antioxidant and anti-arthritic potential of zingiber officinale Rosc. by in vitro and in silico analysis. S Afr J Bot 130:45–53
Naidoo D et al (2020) Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: an in silico analysis. J Biomol Struct Dyn 39:1–13
Ngane A, Ngono R et al (2011) Ethnobotanical survey of some cameroonian plants used for treatment of viral diseases. Afr J Plant Sci 5(1):15–21
Nisius B, Sha F, Gohlke H (2012) Structure-based computational analysis of protein binding sites for function and druggability prediction. J Biotechnol 159(3):123–134
Nugroho A et al (2012) In vivo sedative and gastroprotective activities of Salvia plebeia extract and its composition of polyphenols. Arch Pharmacal Res 35(8):1403–1411
Nurton J (2020) Drug repurposing and the COVID-19 pandemic, WIPO Magazine. https://www.wipo.int/wipo_magazine/en/2020/02/article_0004.html
Patil R et al (2010) “Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing” ed Sridhar Hannenhalli. PLoS ONE 5(8):e12029
Pires DEV, Blundell TL, Ascher DB (2015) PkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med cinal Chem 58(9):4066–72. http://structure.bioc.cam.ac.uk/. Accessed 28 Jul 2020
Pollastri MP (2010) Overview on the rule of five. Curr Protoc Pharmacol 49(1):9.12.1-9.12.8
Prasanth DSNBK et al (2020) In silico identification of potential inhibitors from cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1779129
Pushpakom S et al (2018) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discovery 18(1):41–58
Ranney ML, Griffeth V, Jha AK (2020) Critical supply shortages—the need for ventilators and personal protective equipment during the Covid-19 pandemic. N Engl J Med 382(18):E41
Ren DB et al (2014) Separation of nine compounds from Salvia plebeia R.Br. using two-step high-speed counter-current chromatography with different elution modes. J Sep Sci 37(16):2118–2125
Schüttelkopf AW, Van Aalten DMF (2004) “PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D Biol Crystallogr 60(8):1355–1363. http://scripts.iucr.org/cgi-bin/paper?S0907444904011679. Accessed 29 Jul 2020
Shineman DW et al (2014) Overcoming obstacles to repurposing for neurodegenerative disease. Ann Clin Transl Neurol 1(7):512–518
Talevi A, Bellera CL (2020) Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opin Drug Discov 15(4):397–401
Tao J et al (2007) In vitro anti-HIV and -HSV activity and safety of sodium rutin sulfate as a microbicide candidate. Antiviral Res 75(3):227–233
Veeresham C (2012) Natural products derived from plants as a source of drugs. J Adv Pharm Technol Res 3(4):200–201
Wang J et al (2019) Biosynthesis, chemistry, and pharmacology of polyphenols from Chinese salvia species: a review. Molecules 24(1):1–23
Xiu S et al (2020) Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. https://doi.org/10.1021/acs.jmedchem.0c00502
Zhang MQ, Wilkinson B (2007) Drug discovery beyond the ‘rule-of-five.’ Curr Opin Biotechnol 18(6):478–488
Author Information
Department of Biotechnology, M S Ramaiah Institute of Technology, Bengaluru, India