Comparative study between antiviral drugs and natural compounds against “Cryo-EM structure of the SARS-CoV-2 Omicron spike”
Research Articles | Published: 10 August, 2023
First Page: 596
Last Page: 605
Views: 3199
Keywords: Chemo-informatics, Omicron spike, Pharmacokinetics, SAR, UCSF Chimera
Abstract
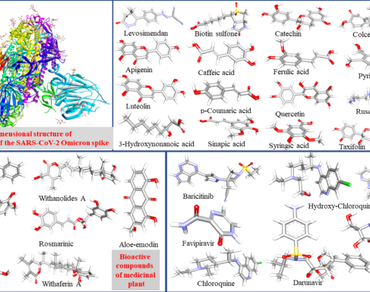
Coronavirus is the + ve sense and single-stranded RNA virus, which mutated continually resulting in new variants such as alpha, delta and Omicron. The available vaccines are not completely effective; however, other oral medications are available against Omicron. Moreover, food products would be helpful to prevent and overcome the complexity of this life-threatening disease. This in-silico study has performed the structural analysis of the “Cryo-EM structure of the SARS-CoV-2 Omicron spike” protein through UCSF Chimera and BIOVIA Discovery Studio. Further, molecular docking was performed to establish the structure-activity relationships (SAR) between the binding pockets of the receptor protein and the natural compounds. SwissADME predicted the pharmacokinetics and drug likeliness of the potential candidates. This study has analyzed the 3 polypeptide chains A, B, C, and 21 heteroatoms present in the 3-D structure of the “Cryo-EM structure of the SARS-CoV-2 Omicron spike” protein. A total of 28 pockets have been found in 7T9J protein by the PrankWeb server. Chain A has 7 ligand binding sites on the peptide group of ASN61, ASN122, ASN165, ASN234, ASN282, ASN331, and ASN343. Molecular docking has predicted that Aloe-emodin (bioactive compounds of Aloe vera) has a good binding affinity (-3.6 kcal/mol) among all the selected natural as well as widely used antiviral compounds. It was found that Sinapic acid (bioactive compounds of pearl millet) also has a good binding affinity (-2.8 kcal/mol) with this Omicron receptor peptide. CABS-flex V2.0 server was used to evaluate the root-mean-square fluctuations (RMSFs) and measure protein stability. The Normal Mode Analysis (Molecular Dynamics simulation) was carried out through iMODs for the validation of the Aloe-emodin and receptor-docked complex. A pharmacokinetic study of predicted compounds favors it to be a good bioactive compound for oral administration. This computational study has revealed the pharmacological relevance of Aloe-emodin and Sinapic acid as potential inhibitors against this deadly virus.
References
Agoni C, Olotu FA, Ramharack P, Soliman ME (2020) Druggability and drug-likeness concepts in drug design: are biomodelling and predictive tools having their say? J Mol Model 26(6). https://doi.org/10.1007/s00894-020-04385-6
Burki TK (2022) Omicron variant and booster COVID-19 vaccines. The Lancet Respiratory Medicine 10(2):e17. https://doi.org/10.1016/S2213-2600(21)00559-2
Cui Z, Liu P, Wang N, Wang L, Fan K, Zhu Q, Wang K, Chen R, Feng R, Jia Z, Yang M, Xu G, Zhu B, Fu W, Chu T, Feng L, Wang Y, Pei X, Yang P, …, Wang X (2022) Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185(5):860–871e13. https://doi.org/10.1016/j.cell.2022.01.019
Diaza RG, Manganelli S, Esposito A, Roncaglioni A, Manganaro A, Benfenati E (2015) Comparison of in silico tools for evaluating rat oral acute toxicity. SAR QSAR Environ Res 26(1):1–27. https://doi.org/10.1080/1062936X.2014.977819
Dyer O (2021) Covid-19: Omicron is causing more infections but fewer hospital admissions than delta, south african data show. BMJ (Clinical Research Ed) 375(November):n3104. https://doi.org/10.1136/bmj.n3104
Eberhardt J, Santos-Martins D, Tillack AF, Forli S (2021) AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python Bindings. J Chem Inf Model 61(8):3891–3898. https://doi.org/10.1021/acs.jcim.1c00203
Fan Y, Li X, Zhang L (2022) SARS-CoV-2 Omicron variant: recent progress and future perspectives. April. https://doi.org/10.1038/s41392-022-00997-x
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD (2021) Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host and Microbe 29(1):44–57e9. https://doi.org/10.1016/j.chom.2020.11.007
Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM (2020) Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 mpro enzyme through in silico approach. Life Sci 255:117831. https://doi.org/10.1016/j.lfs.2020.117831
Halberstein RA (2005) Medicinal plants: historical and cross-cultural usage patterns. Ann Epidemiol 15(9):686–699. https://doi.org/10.1016/j.annepidem.2005.02.004
Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7):409–424. https://doi.org/10.1038/s41579-021-00573-0
Hetz C, Zhang K, Kaufman RJ (2020) Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21(8):421–438. https://doi.org/10.1038/s41580-020-0250-z
Karim SSA, Karim QA (2021) Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. The Lancet 398(10317):2126–2128. https://doi.org/10.1016/S0140-6736(21)02758-6
Khalifa SAM, Mohamed BS, Elashal MH, Du M, Guo Z, Zhao C, Musharraf SG, Boskabady MH, El-Seedi HHR, Efferth T, El-Seedi HR (2020) Comprehensive overview on multiple strategies fighting covid-19. Int J Environ Res Public Health 17(16):1–13. https://doi.org/10.3390/ijerph17165813
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE (2021) PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res 49(D1):D1388–D1395. https://doi.org/10.1093/nar/gkaa971
López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P (2014) IMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res 42(W1):271–276. https://doi.org/10.1093/nar/gku339
Mani JS, Johnson JB, Steel JC, Broszczak DA, Neilsen PM, Walsh KB, Naiker M (2020) Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res 284:197989. https://doi.org/10.1016/j.virusres.2020.197989
Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS, Marquez AC, Sekirov I, Subramaniam S (2022) SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 375(6582):760–764. https://doi.org/10.1126/science.abn7760
May A, Sieker F, Zacharias M (2008) How to efficiently include receptor flexibility during computational docking. Curr Comput Aided-Drug Des 4(2):143–153. https://doi.org/10.2174/157340908784533265
Mukherjee I, Chakrabarti S (2021) Co-evolutionary landscape at the interface and non-interface regions of protein-protein interaction complexes. Comput Struct Biotechnol J 19:3779–3795. https://doi.org/10.1016/j.csbj.2021.06.039
Murgueitio MS, Bermudez M, Mortier J, Wolber G (2012) In silico virtual screening approaches for anti-viral drug discovery. Drug Discovery Today: Technologies 9(3):219–225. https://doi.org/10.1016/j.ddtec.2012.07.009
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel Journal of Cheminformatics 3(33):1–14. https://jcheminf.biomedcentral.com/track/pdf/https://doi.org/10.1186/1758-2946-3-33
Plewczynski D, Hoffmann M, Von Grotthuss M, Ginalski K, Rychewski L (2007) In silico prediction of SARS protease inhibitors by virtual high throughput screening. Chem Biology Drug Des 69(4):269–279. https://doi.org/10.1111/j.1747-0285.2007.00475.x
Rabi FA, Zoubi A, Al-Nasser MS, Kasasbeh AD, G. A., Salameh DM (2020) Sars-cov-2 and coronavirus disease 2019: what we know so far. Pathogens 9(3):1–14. https://doi.org/10.3390/pathogens9030231
Raj U, Varadwaj PK (2016) Flavonoids as multi-target inhibitors for Proteins Associated with Ebola Virus: in Silico Discovery using virtual screening and molecular Docking Studies. Interdisciplinary Sci – Comput Life Sci 8(2):132–141. https://doi.org/10.1007/s12539-015-0109-8
Raschka S, Wolf AJ, Bemister-Buffington J, Kuhn LA (2018) Protein–ligand interfaces are polarized: discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J Comput Aided Mol Des 32(4):511–528. https://doi.org/10.1007/s10822-018-0105-2
Sayed AM, Khattab AR, AboulMagd AM, Hassan HM, Rateb ME, Zaid H, Abdelmohsen UR (2020) Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale. RSC Adv 10(34):19790–19802. https://doi.org/10.1039/d0ra04199h
Sirois S, Wei DQ, Du Q, Chou KC (2004) Virtual screening for SARS-CoV protease based on KZ7088 pharmacophore points. J Chem Inf Comput Sci 44(3):1111–1122. https://doi.org/10.1021/ci034270n
Sun Z, Liu Q, Qu G, Feng Y, Reetz MT (2019) Utility of B-Factors in protein science: interpreting rigidity, flexibility, and Internal Motion and Engineering thermostability [Review-article]. Chem Rev. https://doi.org/10.1021/acs.chemrev.8b00290
Vidal-Limon A, Aguilar-Toalá JE, Liceaga AM (2022) Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for studying Food Proteins and bioactive peptides. J Agric Food Chem 70(4):934–943. https://doi.org/10.1021/acs.jafc.1c06110
Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, …, Ferguson NM (2021) Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593(7858):266–269. https://doi.org/10.1038/s41586-021-03470-x
Zakaryan H, Arabyan E, Oo A, Zandi K (2017) Flavonoids: promising natural compounds against viral infections. Arch Virol 162(9):2539–2551. https://doi.org/10.1007/s00705-017-3417-y
Zhang XQ, Xu X, Bertrand N, Pridgen E, Swami A, Farokhzad OC (2012) Interactions of nanomaterials and biological systems: implications to personalized nanomedicine. Adv Drug Deliv Rev 64(13):1363–1384. https://doi.org/10.1016/j.addr.2012.08.005
Zhao C, Montaseri MH, Wood GS, Pu SH, Seshia AA, Kraft M (2016) A review on coupled MEMS resonators for sensing applications utilizing mode localization. Sens Actuators A 249:93–111. https://doi.org/10.1016/j.sna.2016.07.015
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schön A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, …, Kwong PD (2020) Cryo-EM Structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-Dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host and Microbe 28(6):867–879e5. https://doi.org/10.1016/j.chom.2020.11.004
Kmiecik S, Gront D, Kolinski M, Wieteska L, Dawid AE, Kolinski A (2016) Coarse-grained protein models and their applications. Chem Rev 116:7898–7936
Sumera A, Waseem F, Fatima M, Malik A, Ali N, Zahid A, S (2022) Molecular Docking and Molecular Dynamics Studies reveal secretory proteins as novel targets of Temozolomide in Glioblastoma Multiforme. Molecules 27:7198. https://doi.org/10.3390/molecules27217198
Fatriansyah JF, Rizqillah RK, Yandi MY, Fadilah, Sahlan M (2022) Molecular docking and dynamics studies on propolis sulabiroin-A as a potential inhibitor of SARS-CoV-2. J King Saud Univ - Sci 34(1):101707. https://doi.org/10.1016/J.JKSUS.2021.101707
Pandit A, Shukla AK, Deepika, Vaidya D, Kumari A, Kumar A (2023) In vitro Assessment of Anti-Microbial Activity of Aloe vera (Barbadensis miller) supported through computational studies. Russ J Bioorg Chem. https://doi.org/10.1134/S1068162023020188
Author Information
Department of Nutrition Biology, Central University of Haryana, Mahendergarh, Jant-Pali, India