Clustering based on morphological marker through linear discriminant analysis targeting potent yield enhancers in mini-core collections of chickpea (Cicer arietinum L.)
Research Articles | Published: 18 September, 2023
First Page: 2330
Last Page: 2338
Views: 3478
Keywords: Chickpea, Linear discriminant analysis, Morphological marker, Clustering, Seed yield
Abstract
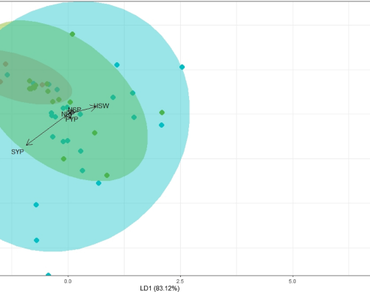
Mini core collections of chickpea, which represent the entire spectrum of diversity were evaluated and discriminated based on the morphological marker, corolla colour. With this objective, 71 accessions were subjected to discriminant function analysis and principal component analysis. From the box’s M test and normal Q–Q plot, the curve was normally distributed and the percentage of separation achieved was 83.12%, 9.54% and 7.34% for LD1, LD2 and LD3, respectively. Confusion matrix indicated that the accuracy level was 72.9% and 56.5% for the training and testing data sets, respectively. The results indicated that corolla colour might not be an effective parameter in discriminating chickpea accessions. From PCA, three components contributed to 72.41% of variability with the maximum PC 1 (45.69%). Factor analysis disclosed that the first two factors contributed to 80% of the variation and the first one is termed as ‘yield predictors’ and the second factor as ‘morphotype predictors’. The accessions ICC4567 and ICC2884 deserve due attention for enhancing yield while ICC4182 could be adjudged as a yield enhancer as well as a superior material for chickpea ideotype breeding. The communalities for all the traits were high [0.9967 (Days to 50% flowering) to 0.6720 (Pod yield per plant)] and indicated the reliability of the investigated variables. The outcomes of the current research adjudged that due weight had to be given during selection upon the mentioned traits and accessions in chickpea yield enhancing projects.
References
Adebisi MA, Okelola FS, Ajala MO, Kehinde TO, Daniel IO, Ajani OO (2013) Evaluation of variations in seed vigour characters of west African rice (Oryza sativa L.) genotypes using multivariate technique. Am J Plant Sci. https://doi.org/10.4236/ajps.2013.42047
AICRP (2020) Project Coordinator’s Report 2019–2020. All India Coordinated Research Project on chickpea. ICAR-Indian Institute of Pulses Research, Kanpur
Akhtar S, Ahmed JU, Hamid A, Islam MR (2010) Evaluation of chickpea (Cicer arietinum L.) genotypes for quality seedlings. The Agriculturists 8(2):108–116. https://doi.org/10.3329/agric.v8i2.7584
Boedeker P, Kearns NT (2019) Linear discriminant analysis for prediction of group membership: a user-friendly primer. Adv Methods Pract Psychol Sci 2:250–263
Chinnasamy K, Arumugam Y, Jegadeesan R, Chockalingam V (2021) Linear discriminant analysis in red sorghum using artificial intelligence. The Nucleus 64(1):103–113. https://doi.org/10.1007/s13237-020-00340-1
Cowen NM, Frey KJ (1987) Relationships between three measures of genetic distance and breeding methods in oat (Avena sativa L.). Genome 29:97–106
Crovello TJ (1970) Analysis of character variation in ecology and systematics. Annu Rev Ecol Syst 1(1):55–98
Deepika C, Nithya V, Vanitha M, Thiruvengadam V (2021) Genetic evaluation of selected accessions of ICRISAT chickpea (Cicer arietinum L.) mini-core collection for seed yield and its component traits. J Food Legumes 34(2):85–91
Denis JC, Adams MW (1978) A factor analysis of the plant variables related to yield in dry beans; Morphological traits. Crop Sci 18:74–78
Dhakar MK, Das B, Nath V, Sarkar PK, Singh AK (2019) Genotypic diversity for fruit characteristics in bael [Aegle marmelos (L.) Corr.] based on principal component analysis. Genet Resour Crop Evol 66(4):951–964
Farshadfar M, Farshadfar E (2008) Genetic variability and path analysis of chickpea (Cicer arientinum L.) landraces and lines. J Appl Sci 8:3951–3956
Fisher RA (1936) The use of multiple measurements in taxonomic problems. Ann Eugenics 7:179–188
Gaur PM, Gour VK (2001) A gene inhibiting flower colour in chickpea (Cicer arietinum L.). Indian J Genet 62(3):273–274
Gaur PM, Thudi M, Samineni S, Varshney RK (2014) Advances in chickpea genomics in legumes in the omic era. In: Gupta S, Nadarajan N, Gupta D (eds) Springer, Berlin, pp 73–94, https://doi.org/10.1007/978-1-4614-83 70–0_4
Gazeli O, Bellou E, Stefas D, Couris S (2020) Laser-based classification of olive oils assisted by machine learning. Food Chem 302:125329. https://doi.org/10.1016/j.foodchem.2019.125329
Gediya LN, Patel DA, Kumar S, Kumar D, Parmar DJ, Patel SS (2019) Phenotypic variability, path analysis and molecular diversity analysis in chickpea (Cicer arietinum L.). Vegetos 32(2):167–180. https://doi.org/10.1007/s42535-019-00020-9
Hasan MT, Deb AC (2017) Assessment of genetic variability, heritability, character association and selection indexes in chickpea (Cicer arietinum L.). Int J Biosci 10(2):111–129. https://doi.org/10.12692/ijb/10.2.111-129
IBPGR, ICRISAT, ICARDA (1993) Descriptors for chickpea (Cicer arietinum L.). IBPGR, ICRISAT and ICARDA
Jain SK, Sharma LD, Gupta KC, Kumar V, Sharma RS (2021) Principal component and genetic diversity analysis for seed yield and its related components in the genotypes of Chickpea (Cicer arietinum L.). Legume Res 1:1–5
Jordaan H, Grove B (2007) Factors affecting maize producers adoption of forward pricing in price risk management: case Vaal. Agrekon 46(4):548–565
Khan AR, Akhtar AR (1934) The inheritance of petal colour in gram. Agric Livestock India 4:127–155
Kumar J (1997) Complementation for flower colour in two chickpea crosses. Indian J Pulses Res 10:227–228
Kumar J, Vijayalakshmi NV, Rao TN (2000) Brief communication. Inheritance of flower color in chickpea. J Heredity 91(5):416–417. https://doi.org/10.1093/jhered/91.5.416
Kumar S, Arora PP, Jeena AS (2003) Correlation studies for yield and its component in chickpea. Agric Sci Digest 23:229–230
Kumar J, Devi M, Verma D, Malik DP, Sharma A (2021) Pre-harvest forecast of rice yield based on meteorological parameters using discriminant function analysis. J Agric Food Res 5:100194. https://doi.org/10.1016/j.jafr.2021.100194
Mansfeld (2008) Cicer arietinum subsp. arietinum. Mansfeld’s world database of agricultural and horticultural crops. IPK Gatersleben. BOTNAME: Cicer %20 arietinum. http://mansfeld.ipk-gatersleben.de/apex/f?p=185:145:::NO::P3_. Accessed 10 Apr 2017
Merga B, Haji J (2019) Economic importance of chickpea: production, value, and world trade. Cogent Food and Agriculture 5:1615718
Pundir RP, Rao NK, Vander Maesen LJG (1985) Distribution of qualitative traits in the world germplasm of chickpea (Cicer arietinum L.). Euphytica 34:697–703. https://doi.org/10.1007/BF00035406
Rajani K, Anand K, Ravi RK, Sadia P, Kumar S, Kumar A, Satyendra Kumar M (2020) Principal component analysis in desi Chickpea (Cicer arietinum L.) under normal sown condition of Bihar. Curr J Appl Sci Technol 39:75–80
Rathore PS, Sharma SK (2003) Scientifc pulse production. Yash Publishing House, Bikaner, p 92
Ribeiro JP, de Medeiros AD, Caliari IP, Trancoso AC, de Miranda RM, de Freitas FC, da Silva LJ, dos Santos Dias DC (2021) FT-NIR and linear discriminant analysis to classify chickpea seeds produced with harvest aid chemicals. Food Chem 342:128324. https://doi.org/10.1016/j.foodchem.2020.128324
Sarker N, Samad MA, Azad AK, Deb AC (2013) Selection for better attributes through variability and discriminant function analysis in chickpea (Cicer arietinum L.). J Subtrop Agric Res Dev 11(1):1050–1055
Sau S, Ucchesu M, D’hallewin G, Bacchetta G (2019) Potential use of seed morpho-colourimetric analysis for Sardinian apple cultivar characterisation. Comput Electron Agric 162:373–379. https://doi.org/10.1016/j.compag.2019.04.027
Sharifi P, Astereki H, Pouresmael M (2018) Evaluation of variations in chickpea (Cicer arietinum L.) yield and yield components by multivariate technique. Ann Agric Sci 16(2):136–142
Sisodia PS, Yasin M (2009) Identification of promising genotypes of Kabuli chickpea (Cicer arietinum L.). Legum Res 32(4):309–310
Swamy SG, Raja DS, Wesley BJ (2020) Susceptibility of stored chickpeas to bruchid infestation as influenced by physico-chemical traits of the grains. J Stored Prod Res 87:101583. https://doi.org/10.1016/j.jspr.2020.101583
Toker C, Cagirgan MI (2004) the use of phenotypic correlations and factor analysis in determining characters for grain yield selection in chickpea (Cicer arietinum L.). Hereditas 140:226–228
Upadhyaya HD (2003) Geographical patterns of variation for morphological and agronomic characteristics in the chickpea germplasm collection. Euphytica 132:343–352. https://doi.org/10.1023/A:1025078703640
Upadhyaya HD, Bramel PJ, Singh S (2001) Development of a chickpea core collection using geographic distribution and quantitative traits. Crop Sci 41:206–210. https://doi.org/10.2135/cropsci2001.411206x
Upadhyaya HD, Bramel PJ, Ortiz R, Singh S (2002) Developing a mini core of peanut for utilization of genetic resources. Crop Sci 42:2150–2156. https://doi.org/10.2135/cropsci2002.2150
Upadhyaya HD, Reddy LJ, Gowda CLL, Reddy KN, Singh S (2006) Development of a mini core subset for enhanced and diversified utilization of Pigeonpea germplasm resources. Crop Sci 46:2127–2132. https://doi.org/10.2135/cropsci2006.01.0032
Upadhyaya HD, Pundir RPS, Dwivedi SL, Gowda CLL, Gopal Reddy V, Singh S (2009) Developing a mini core collection of sorghum [Sorghum bicolor (L.) Moench] for diversified utilization of germplasm. Crop Sci 49:1769–1780. https://doi.org/10.2135/cropsci2009.01.0014
Upadhyaya HD, Sarma NDRK, Ravishankar CR, Albrecht T, Narasimhudu Y, Singh SK, Varshney SK, Reddy VG, Singh S, Dwivedi SL, Wanyera N, Oduori COA, Mgonja MA, Kisandu DB, Parzies HK, Gowda CLL (2010) Developing mini core collection in finger millet using multilocation data. Crop Sci 50:1924–1931. https://doi.org/10.2135/cropsci2009.11.0689
Upadhyaya HD, Ravishankar CR, Narasimhudu Y, Sarma NDRK, Singh SK, Varshney SK, Reddy VG, Singh S, Parzies HK, Dwivedi SL, Nadaf HL, Sharawat KL, Gowda CLL (2011a) Identification of trait-specific germplasm and developing a mini core collection for efficient use of foxtail millet genetic resources in crop improvement. Field Crops Res 124(3):459–467. https://doi.org/10.1016/j.fcr.2011.08.004
Upadhyaya HD, Yadav D, Reddy KN, Gowda CLL, Singh S (2011b) Development of Pearl Millet mini core collection for enhanced utilization of germplasm. Crop Sci 51:217–223. https://doi.org/10.2135/cropsci2010.06.0336
Upadhyaya HD, Dronavalli N, Gowda CL, Singh S (2012) Mini core collections for enhanced utilization of genetic resources in crop improvement. Indian J Plant Genet Resour 25(1):111–124
Xia C, Yang S, Huang M, Zhu Q, Guo Y, Qin J (2019) Maize seed classification using hyperspectral image coupled with multi-linear discriminant analysis. Infrared Phys Technol 103:103077. https://doi.org/10.1016/j.infrared.2019.103077
Author Information
Department of Genetics and Plant Breeding, Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Coimbatore, India