Classification of black gram [Vigna mungo (L.) Hepper] germplasm based on the genetic differences for yield attributing traits
Barathi M. Bala, Babu D. Ratna, Babu J. Sateesh, Ahammed S. Khayum, Rao V. Srinivasa
Research Articles | Published: 14 December, 2023
First Page: 2627
Last Page: 2635
Views: 3814
Keywords: Black gram, Principal component analysis, Agglomerative hierarchial cluster analysis, Transgressive segregants
Abstract
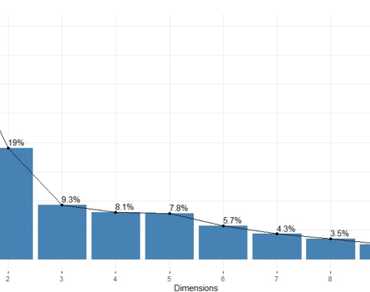
Fifty black gram genotypes were evaluated to identify potential genotypes based on their spatial distances. Principal component analysis (PCA) revealed that first two PC’s recorded more than one eigen value and explained 57.2% of total variation. The principal components PC I and PC II contributed 38.2%, and 19.02%, respectively. The first five components cumulatively explained 82.390% of variation towards total variability. The first principal component (PC-I) contributed maximum by grain yield plant−1, number of pods plant−1, test weight, number of seeds pod−1, pod length and number of branches plant−1. PC-II had maximum variation for days to 50% flowering, days to maturity and plant height. The genotypes were distributed into eight different clusters, while traits got clumped in to two groups in two dimensional (2D) scatter plots. The traits including number of branches plant−1, number of clusters plant−1, number of pods plant−1, number of seeds pod−1, test weight and grain yield plant−1 in first group; days to 50% flowering, days to maturity and plant height under second group were positively correlated among themselves. The traits grain yield plant−1, number of pods plant−1, number of clusters plant−1, test weight, number of branches plant−1, pod length and number of seeds pod−1 had longer vector lengths indicating higher discriminating power in evaluating germplasm. While, the agglomerative hierarchial cluster analysis of genotypes resulted in nine clusters. The clustering pattern in both clustering methods was overlapping for majority of genotypes. The genotypes viz., LBG 932, LBG 787, VBN 11, VBN 8, DKU 87, TBG 141 and GBG 12 were grouped into different clusters in both the methods and can be used in hybridization programmes for isolating transgressive segregants.
References
Al-Naggar AMM, Shafik MM, Musa RYM (2020) Genetic diversity based on morphological traits of 19 maize genotypes using principal component analysis and GT biplot. Ann Res Rev Biol 35(2):68–85
Anderberg MR (1993) Cluster Analysis for Application. Academic Press, New York
Ayesha Md, Babu DR, Rajesh AP, Ahmed MdL, Kumar VM (2020) Principal components of genetic diversity in black gram [Vigna mungo (L.) Hepper]. Pharma Innovat J 10(4):250–253
Bhagwat GV, Joseph J, Antony R (2018) Stability of advanced generation of inter varietal crosses in black gram (Vigna mungo L.) through AMMI analysis. Electron J Plant Breed 9(2):465–475
Bhusal TN, Lal GM, Marker S, Synrem GJ (2016) Discrimination of maize (Zea mays l.) inbreds for morphophysiological and yield traits by D2 statistics and principal component analysis (PCa). Asian J Bio Sci 11(1):77–84
Chang D, Keinan A (2014) Principal component analysis characterizes shared pathogenetics from genome wide association studies. PLOS Comput Biol 9(10):1–14
Devi R, Mittal RK, Sood VK, Sharma PN (2020) Genetic diversity analysis of advanced breeding lines derived from interspecific and intervarietal crosses of black gram based on morphological and molecular markers. Legum Res 43(4):480–487
Federer WT (1956) Augmented (or Hoonuiaku) designs. Hawaiian Planters’ Record LV 2:191–208
Fisher RA, Yates F (1967) Statistical Table for Biological Agricultural and Medical Research. Oliver and Boyd, London, pp 46–63
Gana AS, Shaba SZ, Tsado EK (2013) Principal component analysis of morphological traits in thirty-nine accessions of rice (Oryza sativa L.) grown in a rainfed lowland ecology of Nigeria. J Plant Breed Crop Sci 5(10):120–126
Hefny MM, Ali AA, Byoumi TY, Al-Ashry M, Okasha SA (2017) Classification of genetic diversity for drought tolerance in maize genotypes through principal component analysis. J Agric Sci 62(3):213–227
Jackson JE (1991) A user’s guide to principal components. John Wiley and Sons Inc., New York
Kumar GV, Vanaja M, Abraham B, Jyothi N, Sarkar B (2017) Heterosis and combining ability studies in blackgram (Vigna mungo L. Hepper) under alfisols of SAT region, India. Electron J Plant Breed 8(2):541–547
Legendre, L. and Legendre, P. 1984. Ecologia Numerique. Presses de I’Universite du Quebec 1984;Xv+260p, viii+335.
Ministry of Agriculture, 2020–2021. Government of India. www.indiastat.com. Accessed 2 Apr 2023
Nachimuthu VV, Robin S, Sudhakar D, Raveendran M, Rajeswari S, Manonmani S (2014) Evaluation of rice genetic diversity and variability in a population panel by principal component analysis. Indian J Sci Technol 7(10):1555–1562
Reni YP, Ramana MV, Rajesh AP, Madhavi GB, Prakash KK (2022) Genetic divergence studies in blackgram (Vigna mungo L.) genotypes. Pharma Innovat J 11(2):2153–2158
Sasipriya S, Parimala K, Eswari KB, Balram M (2022) Principal component analysis and metroglyph of variation among sesame (Sesamum indicum L.) genotypes. Electron J Plant Breed 13(4):1–9
Shamim MZ, Pandey A (2019) Simultaneous selection model based evaluation of arsenic tolerance in black gram (Vigna mungo L.) using morphological parameters. Legum Res 42(3):314–319
Sharma JR (1988) Statistical and biometrical techniques in plant breeding. New Age International Publishers
Thirumalai R, Murugan S (2020) Multivariate analysis in black gram (Vigna mungo (L.) hepper) genotypes for mungbean yellow mosaic virus (mymv) resistance. Plant Arch 20(1):2473–2480
Viswanatha KP, Yogeesh LN, Amitha K (2016) Morphological diversity study in horsegram (Macrotyloma uniflorum (Lam.) Verdc) based on principal component analysis (PCA). Electron J Plant Breed 7(3):767–770
Author Information
Department of Genetics and Plant Breeding, Agricultural College, ANGRAU, Bapatla, India