Analysis of genetic divergence and population structure through microsatellite markers in normal and quality protein maize genotypes from NW Himalayan region of India
Research Articles | Published: 25 January, 2020
First Page: 194
Last Page: 202
Views: 3881
Keywords: Maize, SSR markers, Quality protein maize, Population structure
Abstract
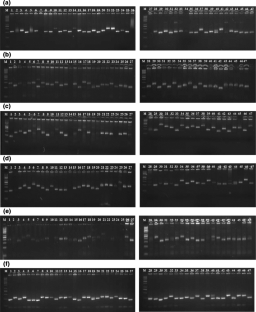
Maize is a self-compatible but largely cross pollinated crop and considered as the staple food in many developing countries. The present study was designed to examine the best polymorphic SSR markers of any maize variety and the assessment of population structure variation and genetic diversity among normal and quality protein maize genotypes. The markers used were found highly polymorphic, that can be effectively used for maize genotypes across the geographical regions. The PIC values for all SSR markers in 47 maize genotypes varied from 0.20 to 0.85 with a mean of 0.57 whereas, gene diversity ranged from 0.23 to 0.86. On the basis of SSR analysis and obtained PIC value (≥ 0.75), gene diversity (≥ 0.79), inbreeding coefficient (≥ 0.97) and polymorphic alleles (≥ 8), six highly polymorphic SSR loci, umc2071, umc1077, umc2163, umc1804, phi026 and umc1799 were observed. The clustering of the maize genotypes was found to be largely based on their centre of development, quality nature and parentage involved. The clustering of the maize genotypes was largely based on their centre of development, quality nature and parentage involved. The genotype CM212 is one of the parent of VQL1 which was converted through MAS, hence clustered with CM212. Likewise, the QPM inbred line VQL373 was derived from the V373 recipient parent through marker assisted conversion, hence clustered together. The finding of this study showed parentage influencing more than the quality related genes. The inferred ancestry at K = 2 suggested that the maize genotypes were grouped into two populations.
References
- Anonymous (2013) Hand book of statistics. Government of India, New Delhi
- ICAR-VPKAS (2012–2013). Annual report. Vivekananda Parvatiya Krishi Anusansthan, Almora.
- Babu BK, Agrawal PK, Mahajan V, Gupta HS (2009) Molecular and biochemical characterization of short duration quality protein maize (QPM). J Plant Biochem Biotech 18(1):93–96
- Babu BK, Pooja P, Bhatt JC, Agrawal PK (2012) Characterization of Indian and exotic quality protein maize (QPM) and normal maize (Zea mays L.) inbreds using simple sequence repeat (SSR) markers. Afr J Biotech 11(41):9691–9700
- Babu BK, Meena V, Agarwal V, Agrawal PK (2014) Population structure and genetic diversity analysis of Indian and exotic rice (Oryza sativa L.) accessions using SSR markers. Mol Biol Rep 41(7):4329–4339
- Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34:20–25
- Enoki H, Sato H, Koinuma K (2002) SSR analysis of genetic diversity among maize inbred lines adapted to cold regions of Japan. Theor Appl Genet 104:1270–1278
- Inghelandt DV, Albrecht EM, Lebreton C, Benjamin S (2010) Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor Appl Genet 120:1289–1299
- Jaccard P (1908) Nouvelles recherches sur la distribuition florale. Bull Soc Vaud Nat 44:223–270
- Kumar B, Rakshit S, Singh RD, Gadag RN, Nath R, Paul AK, Wasialam (2008) Diversity analysis of early maturing elite Indian maize (Zea mays L.) inbred lines using simple sequence repeats. J Plant Biochem Biotechnol 17:133–140
- Legesse BW, Myburg AA, Pixley KV, Botha AM (2007) Genetic diversity of African maize inbred lines revealed by SSR markers. Hereditas 144:10–17
- Liu K, Muse SV (2005) Powermarker: integrated analysis environment for genetic marker data. Bioinformatics 21(9):2128–2129
- Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW (1994) Extraordinarily polymorphic microsatellite dna in barley, species diversity, chromosomal location and population dynamics. Proc Natl Acad Sci USA 91:5466–5470
- Moeller DA, Schaal BA (1999) Genetic relationships among Native American maize accessions of the Great Plains assessed by RAPDs. Theor Appl Genet 99:1061–1067
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
- Pressoir G, Berthaud J (2004) Patterns of population structure in maize landraces from the Central Valleys of Oaxaca in Mexico. Heredity 92:88–94
- Pritchard JK, Wen W (2003) Documentation for the structure software version 2. Department of Human Genetics, University of Chicago, Chicago. https://pritch.bsd.uchicago.edu/software
- Senior ML, Murphy JP, Goodman MM (1998) Utility of SSRs for determining genetic similarities and relationships in maize using agarose gel system. Crop Sci 38:1088–1098
- Smith JSC, Chin ECL, Shu H (1997) An evaluation the utility of SSR loci as molecular markers in maize (Zea mays L.): comparison with data from RFLP and pedigree. Theor Appl Genet 95:163–173
- Uhlig SJ, Bhat BA (1979) Choice of technique in maize milling. Scottish Academic Press Ltd, Edinburgh
- Patto MV, Satovic Z, Pêgo S (2004) Assessing the genetic diversity of Portuguese maize germplasm using microsatellite markers. Euphytica 137:63–72
- Vigouroux Y, Jeffrey CG, Yoshihiro M, Major MG, Jes G, John D (2008) Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Amer J Bot 95(10):1240–1253
- Yao L, Kang F, Guang P (2008) Genetic diversity based on SSR markers in maize (Zea mays L.) landraces from Wuling mountain region in China. J Genet 87:3
Author Information
Department of Agricultural Biotechnology, Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut, India