Comparative study on detection efficacy of universal potyviruses primers
Sharma Abhishek, Sharma Shikha, Khan Irfan, Kaur Sukhjeet, Kaur Sumit Inder
Research Articles | Published: 02 July, 2021
Online ISSN : 2229-4473.
Website:www.vegetosindia.org
Pub Email: contact@vegetosindia.org
First Page: 745
Last Page: 749
Views: 1759
Keywords:
ELISA, Nucleic acid based assay, Vegetables
Abstract
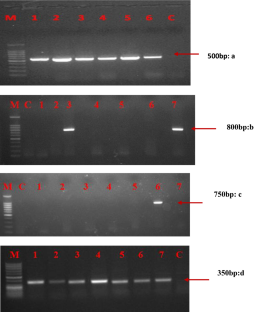
Four universal primer pairs were tested and compared for specificity and efficiency of detection of diverse potyviruses in vegetable crops. Symptomatic samples of seven vegetable crops were initially exposed to DAS/TAS-ELISA. After initial serological detection positive samples were further exposed to different universal primer pairs. All the primers correspond to most conserved regions/gene of potyvirus genome viz., Nuclear Inclusion B and coat protein. Out of four pairs tested, most potential and reproducible candidate universal primer for detection was NIb2F/3R. This set of primer has great breadth of specificity as compared to others, as it was able to detect six different potyviruses. Comparative success to detect potyviruses was maximum with primers pair NIb2F/NIb3R i.e. 84%, 22 and 20% with P9502/CPUP and Sprimer/M4T, respectively.
(*Only SPR Members can get full access. Click Here to Apply and get access)
References
Chen J, Adams MJ (2001) A universal PCR primer to detect members of the Potyviridae and its use to examine the taxonomic status of several members of the family. Arch Virol 146:757–766
Fuji S, Yamamoto H, Furuya H, Naito H (2003) Characterization of a new potyvirus isolated from Chinese artichoke in Japan. Arch Virol 148:2249–2255
Gibbs A, Mackenzie A (1997) A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J Virol Methods 63:9–16
Gibbs AJ, Hajizadeh M, Ohshima K, Jones RA (2020) The potyviruses: an evolutionary synthesis is emerging. Viruses 12:132
Inoue-Nagata AK, Fonseca Sende RO, Boiteux LS, Monte DC, Dusi AN, de Avila AC, van der Vlugt RAA (2002) Pepper yellow mosaic virus, a new potyvirus in sweetpepper, Capsicum annuum. Arch Virol 147:849–855
Jones RAC, Naidu RA (2019) Global dimensions of plant virus diseases: current status and future perspectives. Annu Rev Virol 6:387–409
Kaur S, Kang SS, Sharma A, Sharma S (2014) First report of Pepper mottle virus infecting chilli pepper in India. New Dis Rep 30:2044–0588
Polischuk V, Budzanivska I, Shevchenko T, Oliynik S (2007) Evidence for plant viruses in the region of Argentina Islands, Antarctica. FEMS Microbiol Eecol 59:409–417
Riechmann JL, Lain S, Garcia JA (1992) Highlights and prospects of potyvirus molecular biology. J Gen Virol 73:1–16
Saha A, Saha B, Chakraborty P, Sarkar P, Das S, Saha D (2014) Molecular detection and diversity analysis of some potyviruses associated with mosaic diseases of papaya, common bean and potato growing in sub-Himalayan West Bengal. Vegetos 27:329–337
Sastry KS (2013) Seed-borne plant virus diseases. Springer, New Delhi, pp 10–30
Sharma S, Kang SS, Sharma A (2018) Seed transmissibility of Pepper mottle virus: survival of virus. Curr Sci 115:1–3
Sorel M, Garcia JA, German-Retana S (2014) The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. MPMI 27:215–226
Valouzi H, Golnaraghi A, Hashemi SS, Abedini-Aminabad L, Yazdani-Khameneh S (2019) Detection of potyviruses infecting Iranian common beans using broad-spectrum antibodies and universal primer pairs. Can J Plant Pathol 41:535–543
Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM (2017) ICTV report consortium . J Gen Virol 98:352–354
Zheng L, Rodonibc BC, Gibbs MJ, Gibbs AJ (2010) A novel pair of universal primers for the detection of potyviruses. Plant Pathol 59:211–220
Zheng L, Wayper PJ, Gibbs AJ, Fourment M, Rodoni BC, Gibbs MJ (2008) Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS ONE 3:e1586
Acknowledgements
Author Information
Department of Vegetable Science, College of Horticulture and Forestry, Punjab Agricultural University, Ludhiana, India
Department of Plant Pathology, College of Agriculture, Punjab Agricultural University, Ludhiana, India
shikhasharma-coapp@pau.edu
Department of Plant Pathology, College of Agriculture, Punjab Agricultural University, Ludhiana, India
Department of Vegetable Science, College of Horticulture and Forestry, Punjab Agricultural University, Ludhiana, India
Department of Plant Pathology, College of Agriculture, Punjab Agricultural University, Ludhiana, India